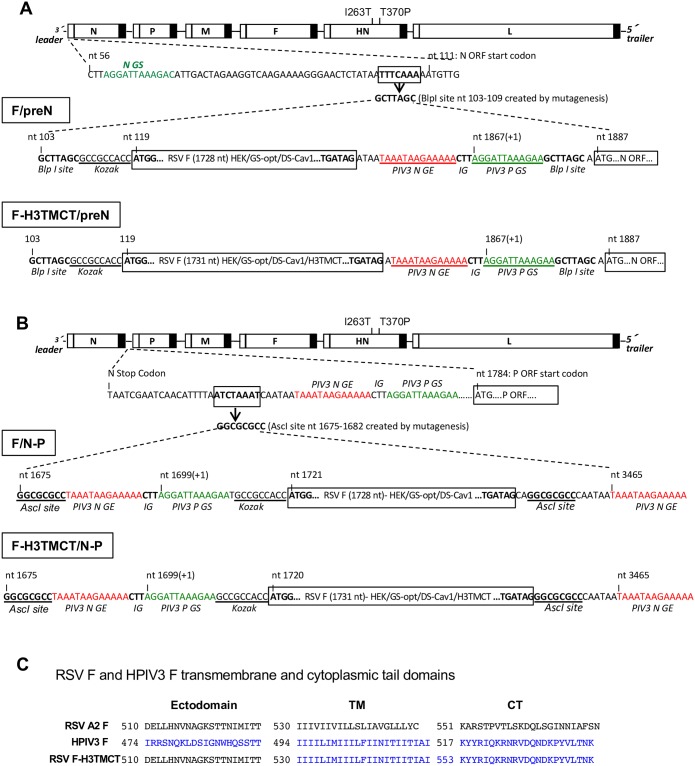
Fig 1. Genome structures of the rHPIV3-RSV-F vectors.
The codon-optimized RSV F ORF containing the HEK and DS-Cav1 mutations (see Results) was engineered to contain an optimal translational start site (underlined and labeled “Kozak” [36]) and was flanked by HPIV3 gene-end (GE) and gene-start (GS) transcription signals and the intergenic CTT trinucleotide to allow for expression as a separate mRNA. RSV F was either the full-length protein (constructs F/preN and F/N-P) or a chimeric version in which the transmembrane (TM) and cytoplasmic tail (CT) domains of RSV F were replaced by those of the HPIV3 F protein (constructs F-H3TMCT/preN and F-H3TMCT/N-P). The insert was placed (A) at the first gene position using the BlpI site or (B) the second gene position using the AscI site in the wild type (wt) rHPIV3 JS strain vector. The vector contained I263T and T370P amino acid substitutions that restore the wt sequence (see the text). (C) Nucleotide sequences of the TM and CT domains and part of the adjoining ectodomains of RSV strain A2 F protein (top), HPIV3 F protein (middle), and chimeric RSV-F-H3TMCT protein.