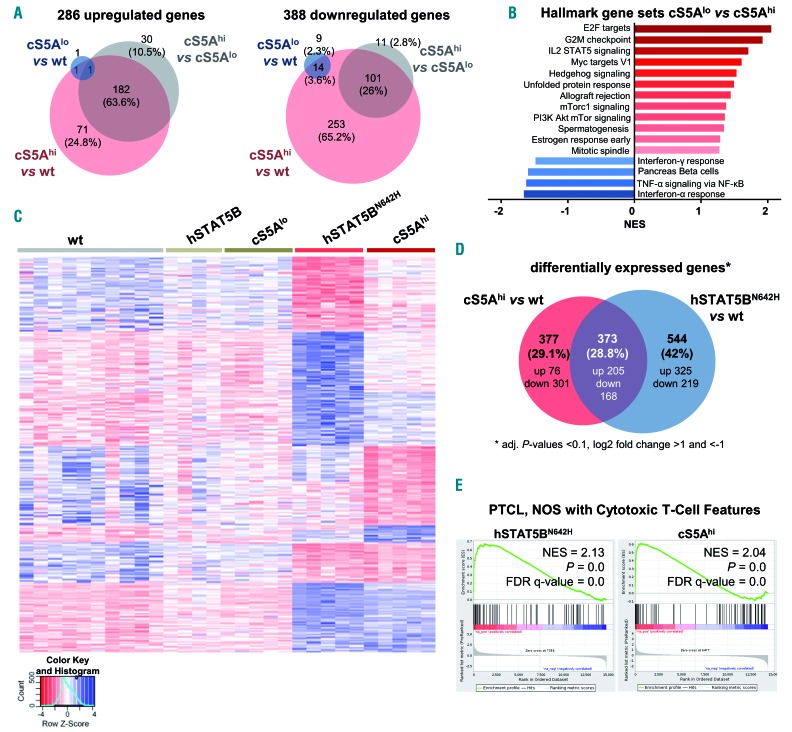
Figure 5.
Transcriptional profiling reveals close correlation to human peripheral T-cell lymphoma. (A) CD8+ T-cell RNA-sequencing (performed with Illumina HiSeq2500) analysis showing the number of significantly (left) up- and (right) down-regulated genes in wildtype (wt), cS5Alo and cS5Ahi CD8+ T cells obtained from lymph nodes (n=5/genotype; adjusted P-values <0.1, log2 fold change >1 and <-1). (B) Summary of gene set enrichment analysis (GSEA) of hallmark gene sets enriched in cS5Ahi vs. cS5Alo CD8+ T cells [false discovery rate (FDR) ≤0.25, adjusted P-value ≤0.05)]. (C) RNA isolated from CD8+ T cells of wt (n=10), hSTAT5B (n=4), hSTAT5BN642H, cS5Alo and cS5Ahi (all n=5) mice were subjected to RNA-sequencing. Heatmap of genes deregulated in a comparison of all genotypes to wt controls (n=1,055) clustered for up- and down-regulated genes specific to individual conditions, as well as for genes shared between hSTAT5BN642H and cS5Ahi. Scaled, rlog transformed normalized counts from DESeq2 were used for the analysis. (D) Venn diagram of differentially expressed genes in cS5Ahi vs. wt and hSTAT5BN642H vs. wt CD8+ T cells (adjusted P-values <0.1, log2 fold change >1 and <-1). (E) GSEA of STAT5BN642H and cS5Ahi expression data shows a correlation to peripheral T-cell lymphoma, not otherwise specified (PTCL, NOS) with cytotoxic T-cell features. NES: normalized enrichment score. The gene set was compiled from literature.6,7