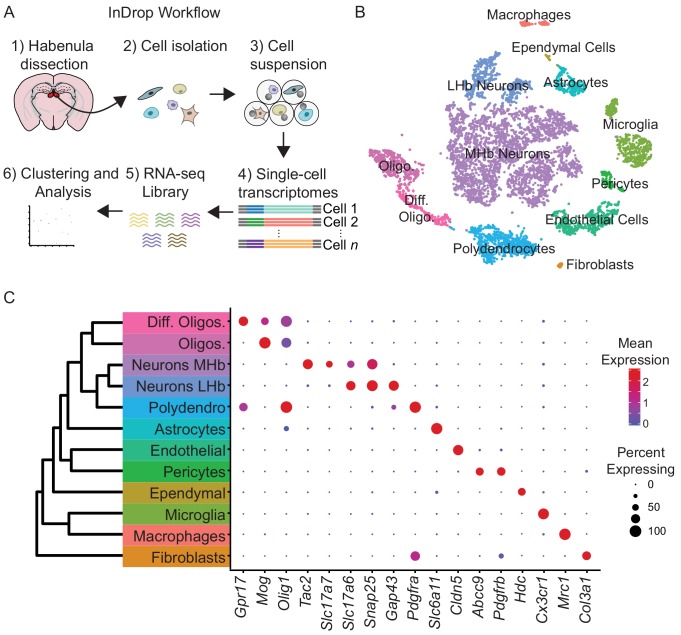
Figure 1. High-throughput single-cell transcriptomic profiling of the habenula.
(A) Schematic for scRNA-seq using the inDrop platform. Tissue containing the habenula was microdissected from acute coronal brain slices prepared from adult mice (1). Tissue chunks were digested in a cocktail of proteases and followed by trituration and filtration to obtain a cell suspension (2). Single cells were encapsulated using a droplet-based microfluidic device (3) for cell barcoding and mRNA capture (4). RNA sequencing (5) and bioinformatics analysis followed (6). (B) t-SNE plot of the processed dataset containing 7,506 cells from six animals. Cells are color-coded according to the cluster labels shown in (C). (C) Left: Dendrogram with cell class labels corresponding to clusters shown in (B). Right: Dot plot displaying expression of example enriched genes used to identify each major cell class. The color of each dot (blue to red) indicates the relative log-scaled expression of each gene, whereas the dot size indicates the fraction of cells expressing the gene.