Abstract
The inhibitor of differentiation/DNA-binding (ID) is a member of the helix–loop–helix (HLH) transcription factor family, and plays a role in tumorigenesis, invasiveness and angiogenesis. The aims were to investigate the expression patterns and prognostic values of individual ID family members in lung cancer, and the potential functional roles. The expression levels of ID family were assessed using the Oncomine online database and GEPIA database. Furthermore, the prognostic value of ID family members was evaluated using the Kaplan–Meier plotter database. The genetic mutations of ID family members were investigated using the cBioPortal database. Moreover, enrichment analysis was performed using STRING database and Funrich software. It was found that all the ID family members were significantly down-regulated in lung cancer. Prognostic results indicated that low mRNA expression levels of ID1 or increased mRNA expression levels of ID2/3/4 were associated with improved overall survival, first progression and post progression survival. Additionally, genetic mutations of ID family members were identified in lung cancer, and it was suggested that amplification and deep deletion were the main mutation types. Furthermore, functional enrichment analysis results suggested that ID1/2/4 were significantly enriched in ‘regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism’ for biological process, ‘transcription factor activity’ for molecular function and ‘HLH domain’ for protein domain. However, it was found that ID3 was not enriched in the above functions. The aberrant expression of ID family members may affect the occurrence and prognosis of lung cancer, and may be related to cell metabolism and transcriptional regulation.
Keywords: expression, inhibitor of differentiation/DNA-binding, lung cancer, prognostic, protein interaction network
Introduction
Lung cancer is one of the most common types of malignancies worldwide, and has higher morbidity and mortality rates compare with other malignant tumors [1]. As a threat to human health, lung cancer has been the focus of the health research field worldwide [2]. Currently, the survival of patients with lung cancer relies on quick and accurate clinical diagnosis. However, most patients have intermediate or advanced lung cancer at the time of diagnosis, and thus have a poor survival rate [3]. Therefore, it is important to investigate potential biomarkers that are related to the occurrence and prognosis of lung cancer, and to identify the possible molecular mechanisms.
The inhibitor of differentiation/DNA-binding (ID) belongs to helix–loop–helix (HLH) family of transcription factors. ID can bind to HLH transcription factors to inhibit their binding to DNA, resulting in the inhibition of cell differentiation and promotion of cell proliferation [4]. In humans, the ID family members consist of four members: ID1, ID2, ID3 and ID4. Previous studies have demonstrated that ID proteins play important roles in proliferation, apoptosis, differentiation, invasion, metastasis and angiogenesis in various human tumor types [5–7]. However, the role of ID family members in lung cancer is not fully understood.
Kamalian et al. [8] found that the expression levels of ID1, ID2 and ID3 were increased in human small cell lung cancer (SCLC) tissues and cell lines, and that only ID4 was increased in SCLC tissues. In addition, Kamalian et al. [8] revealed that the increased expression level of ID2 in the cytoplasm of patients with SCLC is significantly associated with increased survival. Therefore, ID2 may be used as a prognostic factor of patients with SCLC. Cheng et al. also reported that the expression level of ID1 is increased in lung cancer cell lines and tissues, and promotes lung cancer cell proliferation and tumor growth via the Akt-related signaling pathway [9]. Furthermore, elevated expressions levels of ID1 and ID3 are associated with SCLC tumorigenicity by enhancing angiogenesis and suppressing apoptosis [10]. Moreover, Li et al. found that the effects of ID1 in non-SCLC (NSCLC) cells may promote proliferation, migration and invasion by activating the NF-κB signaling pathway [11]. In addition, Qi et al. found that ID4 could inhibit cisplatin-induced apoptosis via the p38 MAPK pathway [12]. In NSCLC, which affects 80% of patients with lung malignancies, ID2 may be involved in dedifferentiation, and could be used as a prognosis marker for patients with poorly differentiated tumors [13]. Therefore, ID family members may provide new targets and biomarkers for the treatment and prognosis of lung cancer.
Bioinformatics analysis has been widely applied in the biology research field. The present study investigated the role of individual ID family members in lung cancer using large databases, which included the Oncomine database, Gene Expression Profiling Interactive Analysis (GEPIA) database, Kaplan–Meier plotter, cBio Cancer Genomics Portal (cBioPortal) database, Search Tool for the Retrieval of Interacting Genes (STRING) database and Funrich software. The aims of the present study were to examine the characteristics of individual ID family members in lung cancer, including the expression levels, prognostic values and potential functions, to further the understanding of ID proteins.
Materials and methods
Oncomine analysis
The Oncomine online database (www.oncomine.org) is an online cancer microarray database, which is used to compare the expression between tumors and normal tissues in various cancer types [14]. The selection criteria included: P < 0.05, 2-fold change and gene rank in the top 10%.
GEPIA database
The GEPIA database (http://gepia.cancer-pku.cn/) is a newly developed interactive web server for comparing the gene expression profile in cancer and paired normal tissues [15]. The present study compared the expression profile of ID family members in lung cancer using the GEPIA database.
Kaplan–Meier analysis
The prognostic values of ID family members in lung cancer were assessed using the Kaplan–Meier plotter (www.kmplot.com), which can evaluate the effect of 54,000 genes on survival rate in 21 cancer types including breast cancer, lung cancer and ovarian cancer [16,17]. A total of 1926 lung cancer samples were used to assess first progression (FP), overall survival (OS) and post progression survival (PPS), using a Kaplan–Meier survival plot. In addition, the present study evaluated the associations of the ID family members with different clinical parameters for OS, including the histology, clinical stages, pathological grades, American Joint Committee on Cancer (AJCC) stages, sex and smoking status. The log-rank P < 0.05 was considered to indicate a statistically significant difference.
cBioPortal database
The cBioPortal database (http://cbioportal.org) is an open-access resource for the interactive investigation of multidimensional cancer genomics datasets [18]. The genetic mutations of ID family members in different cancer types were obtained, according to the online cBioPortal database.
String database
The STRING database (https://string-db.org/) is a search tool for the retrieval of interacting genes, which is used to assess protein–protein interaction (PPI) information [19]. The gene networks of ID family member genes were constructed, and the interactions with a combined score >0.4 were selected.
Funrich analysis
Functional enrichment analysis of the interacting proteins was performed using Funrich software (version 3.1.3), which is an open-access standalone functional enrichment and interaction network analysis tool [20]. Moreover, the functional analysis of related gene interactions with ID family members was performed using Funrich software.
Results
Transcription expression levels of ID family members in human cancer types
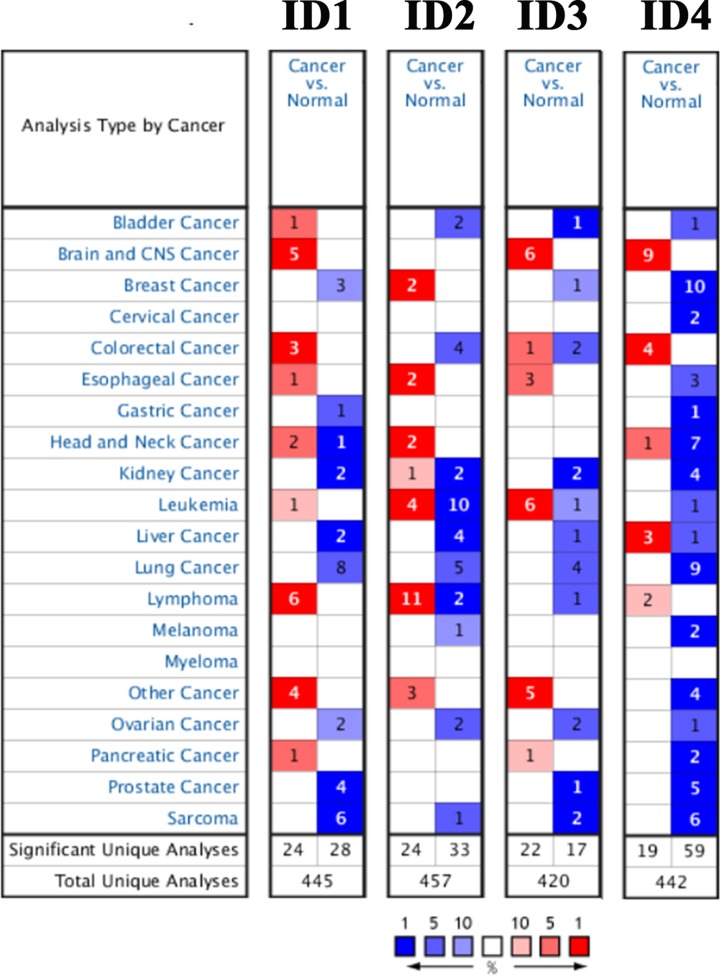
The present study used the Oncomine online databases to compare the transcription expression levels of ID family members between human cancer and normal tissues. As shown in Figure 1, the database containing the genes of ID1, ID2, ID3 and ID4 had a total of 445, 457, 420 and 442 unique studies, respectively. It was found that all ID family members were significantly down-regulated in lung cancer.
Figure 1. mRNA expression levels of ID family genes in the Oncomine database.
The graphic demonstrates the number of datasets with statistical significance. Red, up-regulation; blue, down-regulation. The number in each cell indicates the datasets that met the set threshold in each cancer type. Cell color was defined as the gene rank percentile for analyses within the cell. ID, inhibitor of differentiation/DNA-binding.
Furthermore, the present study investigated the gene expression levels of ID family members in different lung cancer datasets (Table 1). In the Bhattacharjee dataset [21], ID1 was significantly decreased in different lung cancer types compared with normal samples; lung adenocarcinoma had a fold change of -11.038, small cell lung carcinoma had a fold change of -11.150 and lung carcinoma tumor had a fold change of -45.049. Similar results were identified in lung adenocarcinoma in the Beer dataset [22], Su dataset [23], Landi dataset [24], Selamat dataset [25] and Okayama dataset [26]. For ID2, a decreased expression level was found in lung adenocarcinoma compared with normal samples in the Selamat [25], Beer [22], Bhattacharjee [21] and Su [23] datasets. Furthermore, in the Hou dataset [27] it was demonstrated that the expression level of ID2 was decreased in squamous cell lung carcinoma, with a fold change of -2.485. Moreover, a low expression level of ID3 was found in lung adenocarcinoma in the Selamat dataset [25], Landi dataset [24], Su dataset [23] and Okayama dataset [26]. Furthermore, ID4 was significantly down-regulated in lung adenocarcinoma in the Okayama [26], Beer [22], Landi [24], Su [23], Garber [28], Stearman [29] and Hou [27] datasets, and also in squamous cell lung carcinoma (Garber [28] and Hou [27] datasets).
Table 1. The significant change of IDs expression in different types of lung cancer (Oncomine database).
| Dataset | Types of lung cancer versus lung | Sample | Fold change | P-value | t-test | |
|---|---|---|---|---|---|---|
| ID1 | Bhattacharjee | Lung adenocarcinoma versus normal | 149 | -11.038 | 5.25E-7 | -6.259 |
| Small cell lung carcinoma versus normal | 23 | -11.150 | 1.25E-5 | -6.055 | ||
| Lung carcinoid tumor versus normal | 37 | -45.049 | 2.28E-9 | -7.724 | ||
| Beer | Lung adenocarcinoma versus normal | 96 | -5.242 | 1.10E-8 | -6.132 | |
| Su | Lung adenocarcinoma versus normal | 66 | -3.186 | 1.51E-7 | -6.099 | |
| Landi | Lung adenocarcinoma versus normal | 107 | -2.364 | 4.98E-12 | -7.718 | |
| Selamat | Lung adenocarcinoma versus normal | 116 | -3.080 | 9.60E-16 | -9.481 | |
| Okayama | Lung adenocarcinoma versus normal | 246 | -2.652 | 1.57E-9 | -8.345 | |
| ID2 | Selamat | Lung adenocarcinoma versus normal | 116 | -3.864 | 5.22E-27 | -14.089 |
| Beer | Lung adenocarcinoma versus normal | 96 | -2.231 | 3.90E-7 | -6.616 | |
| Bhattacharjee | Lung adenocarcinoma versus normal | 203 | -4.723 | 2.09E-5 | -5.185 | |
| Su | Lung adenocarcinoma versus normal | 66 | -2.050 | 2.14E-7 | -5.852 | |
| Hou | Squamous cell lung carcinoma versus normal | 156 | -2.485 | 2.38E-11 | -9.385 | |
| ID3 | Selamat | Lung adenocarcinoma versus normal | 116 | -4.415 | 1.21E-27 | -14.460 |
| Landi | Lung adenocarcinoma versus normal | 107 | -2.116 | 3.96E-16 | -9.520 | |
| Su | Lung adenocarcinoma versus normal | 66 | -3.906 | 1.50E-6 | -5.575 | |
| Okayama | Lung adenocarcinoma versus normal | 246 | -2.638 | 1.62E-11 | -10.739 | |
| ID4 | Okayama | Lung adenocarcinoma versus normal | 246 | -4.115 | 1.48E-28 | -17.724 |
| Beer | Lung adenocarcinoma versus normal | 96 | -4.988 | 7.24E-10 | -7.656 | |
| Landi | Lung adenocarcinoma versus normal | 107 | -3.754 | 6.51E-25 | -13.467 | |
| Su | Lung Adenocarcinoma versus Normal | 66 | -3.259 | 5.44E-11 | -8.206 | |
| Garber | Lung Adenocarcinoma versus Normal | 45 | -2.495 | 4.65E-6 | -6.296 | |
| Squamous Cell Lung Carcinoma versus Normal | 19 | -3.193 | 7.94E-5 | -4.885 | ||
| Stearman | Lung adenocarcinoma versus normal | 39 | -2.478 | 1.50E-7 | -6.829 | |
| Hou | Lung adenocarcinoma versus normal | 110 | -5.899 | 1.42E-15 | -10.443 | |
| Squamous cell lung carcinoma versus normal | 92 | -7.876 | 3.49E-18 | -14.320 |
Expression levels of ID family members in lung cancer
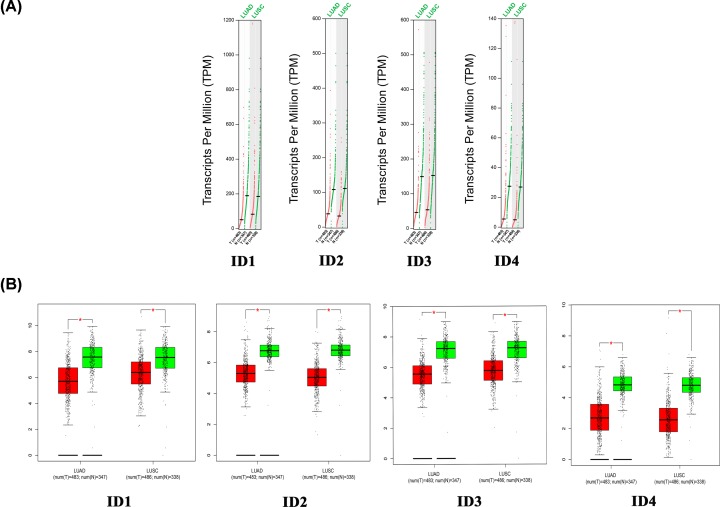
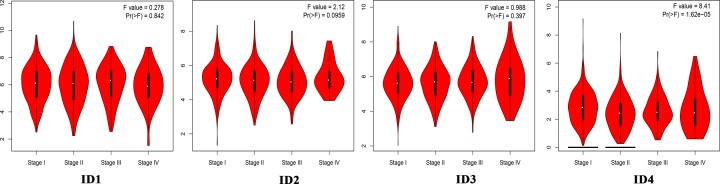
Using the GEPIA database, the present study compared the mRNA expression level of individual ID family members between lung cancer tissues and normal lung tissues. The present results demonstrated that the expression levels of ID family member genes were significantly decreased in lung adenocarcinoma and lung squamous cell carcinoma tissues compared with normal tissues (Figure 2). Furthermore, the present study analyzed the expression levels of ID family members in different lung cancer stages. It was found that ID4 had significantly different expression levels in the various tumor stages, whereas there was no obvious difference in expression levels of ID1, ID2 and ID3 (Figure 3).
Figure 2. Expression of ID family members in lung cancer in the GEPIA database.
(A) Expression profile of ID family members in lung cancer. Red trace, tumor samples; green trace, normal samples. (B) Boxplot results of the expression levels of ID family members in lung cancer. A t-test was used to compare the expression level differences between tumor and normal tissues (P < 0.01). Y-axis represents log2(TPM+1). Red box, tumor samples; green box, normal samples. T, tumor; N, normal; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; ID, inhibitor of differentiation/DNA-binding.
Figure 3. Expression of the ID family members in lung cancer stages.
The Y-axis represents log2(TPM+1). ID, inhibitor of differentiation/DNA-binding.
Prognosis analysis of ID family members in lung cancer
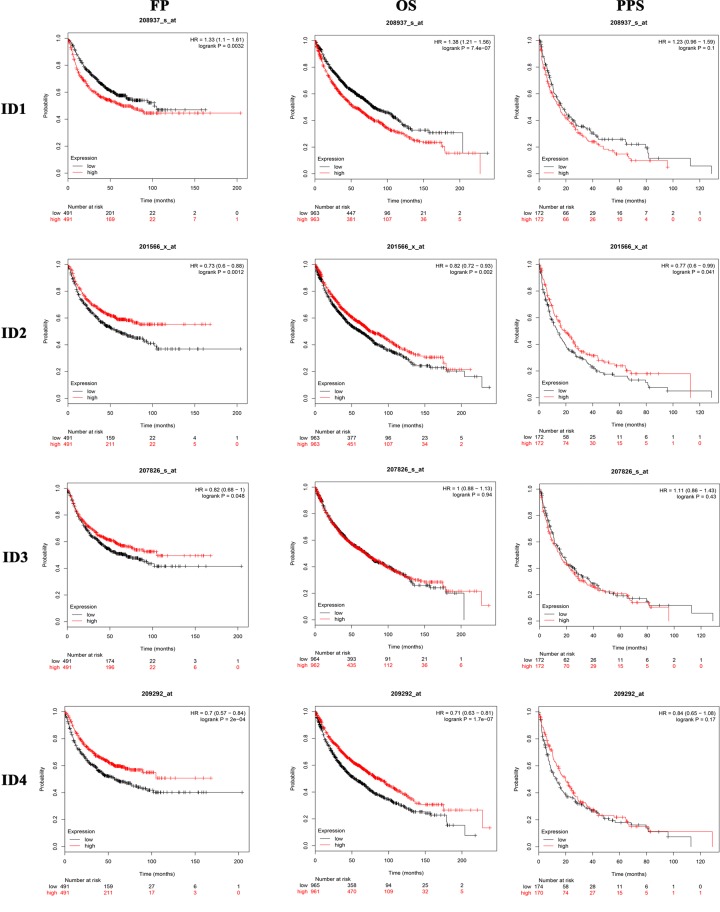
The present study systematically performed Kaplan–Meier survival analysis according to the mRNA expression of individual ID family members in lung cancer (http://www.kmplot.com/analysis/index.php?p=service&cancer=lung). It was found that the mRNA expression levels of ID1/2/3/4 had a significant effect on OS, FP and PPS in patients with lung cancer (P < 0.05; Figure 4). Therefore, patients with lung cancer with a low mRNA expression level of ID1 or high mRNA expression levels of ID2/3/4 were predicted to have longer OS, FP and PPS.
Figure 4. Relationship between the expression level of individual ID family members and the prognosis of patients with lung cancer.
ID, inhibitor of differentiation/DNA-binding; OS, overall survival; FP, progression-free survival; PPS, post-progression survival.
Furthermore, the present study investigates the association of the expression levels of individual ID family members with various clinical parameters of lung cancer, including histology, clinical stages, pathological grades, AJCC stages, sex and smoking status (Table 2). For histology, ID1 mRNA expression level was significantly associated with unfavorable OS in adenocarcinoma and squamous cell carcinoma. However, ID2 and ID4 mRNA expression levels showed a favorable association with OS in adenocarcinoma. For clinical stages, ID1 mRNA expression level was associated with unfavorable OS in patients with stage 1 lung cancer. On the contrary, mRNA expression levels of ID2 and ID4 were related with favorable OS. However, it was found that ID family member mRNA expression levels showed no correlation with stages 2 and 3 of lung cancer. For pathological grades, only ID1 was identified to be associated with unfavorable OS in patients with stage 2 lung cancer. For AJCC stages, it was demonstrated that ID1 (AJCC stage N0), ID2 (AJCC stage M0) and ID3 (AJCC stage T4) were significantly associated with unfavorable OS in lung cancer. However, ID4 (AJCC stage T1) was associated with favorable OS in lung cancer. Moreover, ID1 was significantly correlated with poor OS in female patients with lung cancer, while ID2/3/4 were significantly correlated with improved OS in female patients with lung cancer. In addition, it was found that ID1 was significantly correlated with poor OS in male patients with lung cancer. The present results suggested that ID1 mRNA expression level was significantly associated with poor OS in smokers and non-smokers. However, ID2 and ID4 mRNA expression levels were associated with improved OS in patients with lung cancer without a smoking history.
Table 2. Relation of the expression of individual ID family members with OS in lung cancer patients with different clinical parameters.
| Subtypes | ID1 | ID2 | ID3 | ID4 | ||||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | Log rank P | HR (95% CI) | Log rank P | HR (95% CI) | Log rank P | HR (95% CI) | Log rank P | |
| Histology | ||||||||
| Adenocarcinoma | 1.82(1.43–2.32) | 1.0e-6 | 0.56(0.44–0.71) | 1.1e-6 | 1.15(0.91–1.45) | 0.25 | 0.46(0.36–0.58) | 1.0e-10 |
| Squamous cell carcinoma | 1.4(1.11–1.78) | 0.005 | 1(0.79–1.26) | 0.98 | 0.9(0.71–1.15) | 0.41 | 0.9(0.71–1.14) | 0.4 |
| Stage | ||||||||
| 1 | 2.38(1.78–3.18) | 1.5e-9 | 0.5(0.38–0.67) | 8.3e-7 | 0.98(0.75–1.29) | 0.89 | 0.4(0.3–0.53) | 3.7e-11 |
| 2 | 1.27(0.87–1.83) | 0.21 | 0.91(0.63–1.32) | 0.63 | 0.9(0.63–1.3) | 0.58 | 0.79(0.55–1.14) | 0.21 |
| 3 | 1.19(0.69–2.06) | 0.54 | 1.15(0.67–2) | 0.61 | 1.54(0.89–2.67) | 0.12 | 1.3(0.75–2.24) | 0.35 |
| 4 | NA | NA | NA | NA | NA | NA | NA | NA |
| Grade | ||||||||
| I | 1.4(0.98–2.01) | 0.065 | 0.95(0.66–1.36) | 0.78 | 0.87(0.61–1.25) | 0.46 | 1.06(0.74–1.52) | 0.75 |
| II | 1.46(1.07–2) | 0.017 | 1.25(0.91–1.72) | 0.16 | 0.93(0.68–1.27) | 0.65 | 0.94(0.69–1.29) | 0.72 |
| III | 1.21(0.63–2.33) | 0.57 | 1.02(0.53–1.96) | 0.95 | 0.64(0.33–1.25) | 0.19 | 1.01(0.53–1.95) | 0.97 |
| AJCC stage T | ||||||||
| 1 | 1.23(0.92–1.63) | 0.16 | 0.92(0.69–1.22) | 0.54 | 0.88(0.67–1.17) | 0.39 | 0.67(0.5–0.89) | 0.0059 |
| 2 | 1.18(0.95–1.47) | 0.14 | 1.07(0.86–1.33) | 0.56 | 0.95(0.76–1.18) | 0.62 | 1.07(0.86–1.34) | 0.53 |
| 3 | 1.26(0.77–2.06) | 0.35 | 1.07(0.65–1.77) | 0.79 | 0.85(0.51–1.4) | 0.51 | 0.91(0.56–1.49) | 0.71 |
| 4 | 1.22(0.65–2.29) | 0.53 | 0.91(0.48–1.71) | 0.76 | 2.48(1.27–4.83) | 0.006 | 1.16(0.62–2.18) | 0.65 |
| AJCC stage N | ||||||||
| 0 | 1.31(1.06–1.61) | 0.013 | 0.98(0.79–1.2) | 0.81 | 0.96(0.78–1.19) | 0.71 | 0.91(0.74–1.12) | 0.36 |
| 1 | 1.2(0.88–1.65) | 0.24 | 1.21(0.8–1.65) | 0.24 | 0.88(0.64–1.2) | 0.41 | 0.85(0.62–1.16) | 0.3 |
| 2 | 1.32(0.88–1.98) | 0.18 | 0.94(0.63–1.42) | 0.78 | 1.45(0.96–2.2) | 0.077 | 0.9(0.6–1.35) | 0.61 |
| AJCC stage M | ||||||||
| 0 | 1.22(0.99–1.5) | 0.062 | 1.24(1.01–1.53) | 0.04 | 0.94(0.77–1.16) | 0.57 | 0.84(0.68–1.04) | 0.1 |
| 1 | NA | NA | NA | NA | NA | NA | NA | NA |
| Gender | ||||||||
| Female | 1.27(1–1.6) | 0.047 | 0.74(0.58–0.93) | 0.01 | 0.73(0.57–0.92) | 0.0068 | 0.59(0.47–0.75) | 1.2e-5 |
| Male | 1.45(1.24–1.7) | 4.3e-6 | 0.85(0.73–1) | 0.05 | 1.17(1–1.37) | 0.053 | 0.8(0.69–0.94) | 0.0061 |
| Smoking history | ||||||||
| Exclude those never smoked | 1.55(1.26–1.92) | 3.2e-5 | 0.87(0.71–1.07) | 0.19 | 0.97(0.79–1.19) | 0.79 | 0.92(0.74–1.13) | 0.41 |
| Only those never smoked | 1.81(1.03–3.19) | 0.038 | 0.32(0.17–0.59) | 0.00011 | 0.74(0.42–1.29) | 0.29 | 0.36(0.19–0.67) | 0.00078 |
Mutation of ID family members in lung cancer
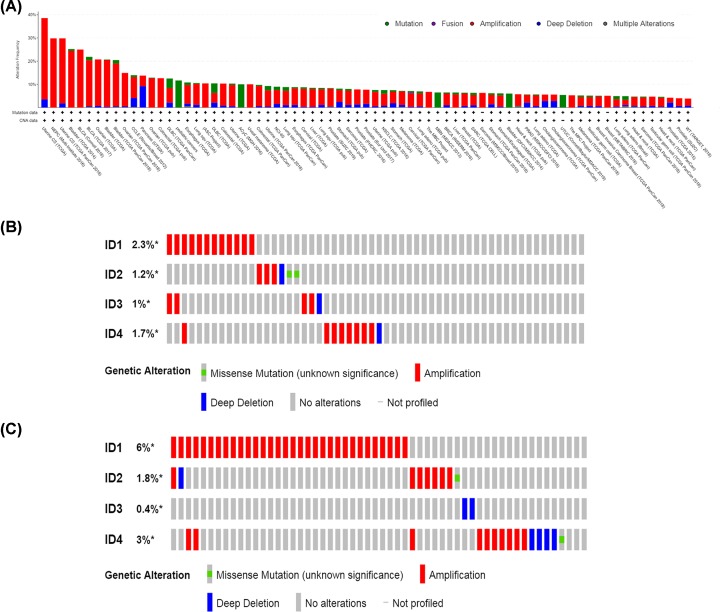
Genetic mutations in ID family members in different cancer types were assessed using cBioPortal. The ID family member genetic mutations were determined in 256 cancer studies, which included 77,879 samples. As shown in Figure 5A, genetic mutations in ID family members were present in different lung cancer types, including lung squamous cell carcinoma and lung adenocarcinoma, compared with other cancer types. Furthermore, ID family genetic mutation frequencies and types in lung adenocarcinoma (TCGA; Provisional; 586 total samples) and lung squamous cell carcinoma (TCGA; Provisional; 511 total samples) are shown in Figure 5B and C. In lung adenocarcinoma (TCGA; Provisional), the mutation frequencies of ID1/2/3/4 were 2.3, 1.2, 1 and 1.7%, respectively, and gene amplification accounted for the majority of genetic modifications. In lung squamous cell carcinoma (TCGA; Provisional), the mutation frequencies of ID1/2/3/4 were 6, 1.8, 0.4 and 3%, respectively, and gene amplification accounted for the majority of genetic changes for ID1/2/4, while deep deletions accounted for the majority of changes for ID3.
Figure 5. Analysis of ID family genetic mutations using the cBioPortal database.
(A) Genetic mutation frequencies of ID family members in various carcinoma types. Green, genetic mutations; purple, gene fusions; red, gene amplifications; blue, deep deletions; gray, multiple alterations. (B) ID family genetic mutation frequencies and mutation types in 584 patients with lung adenocarcinoma. Red, gene amplifications; blue, deep deletions; green, missense mutations. (C) ID family genetic mutation frequencies and mutation types in 511 patients with lung squamous cell carcinoma. Red, gene amplifications; blue, deep deletions; green, missense mutations. ID, inhibitor of differentiation/DNA-binding.
Functional enrichment analysis of ID family members
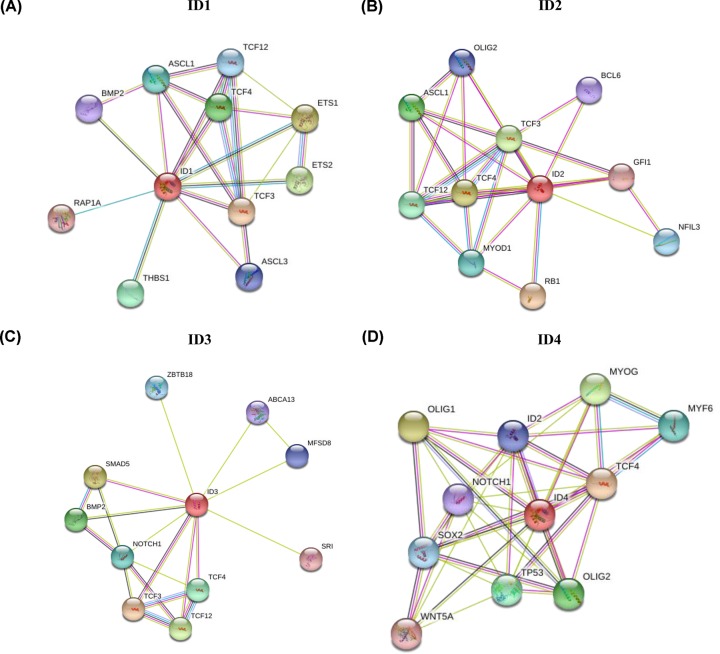
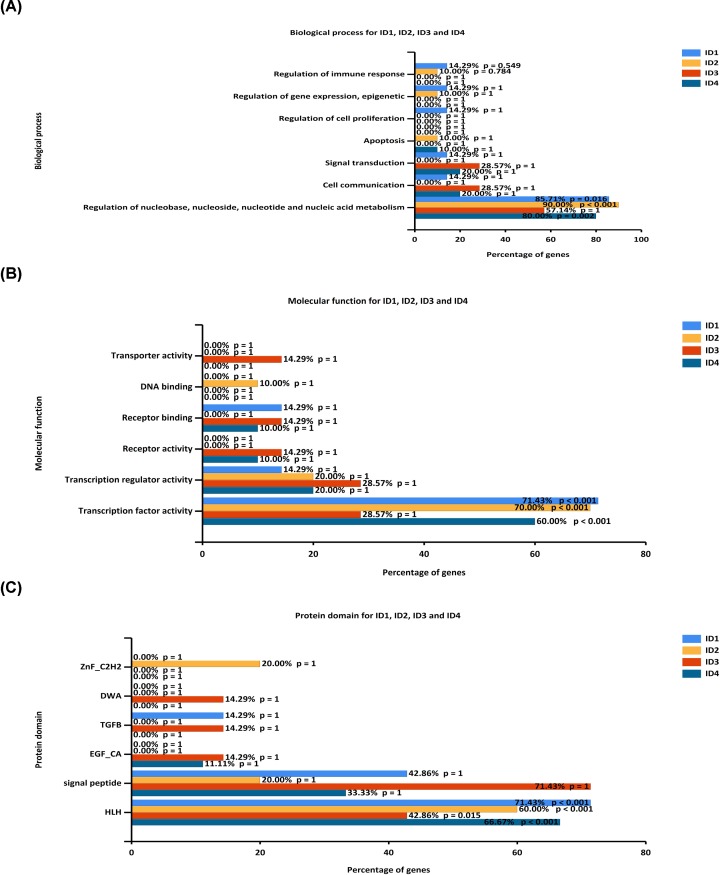
To investigate the interactions of the ID family genes, the present study constructed PPI networks using STRING data (Figure 6). Furthermore, functional enrichment analysis of potential target genes was performed using Funrich software (Figure 7). For biological process, ID1/2/4 were significantly enriched in ‘Regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolis’ (85.71, 90.00 and 80.00%; P<0.05). For molecular function, ID1/2/4 were significantly enriched in ‘Transcription factor activity’ (71.43, 70.00 and 60.00%; P < 0.05). It was found that for protein domain, ID1/2/4 were significantly enriched in ‘HLH domain’ (71.43, 60.00 and 66.67%; P < 0.05). However, ID3 was not significantly enriched in the above aspects.
Figure 6. Protein–protein interaction network of individual ID family members from the STRING database.
(A) Interaction network of ID1. (B) Interaction network of ID2. (C) Interaction network of ID3. (D) Interaction network of ID4. ID, inhibitor of differentiation/DNA-binding.
Figure 7. Functional enrichment analysis of potential targeted genes.
(A) Potential biological processes of ID family genes were identified using FunRich. (B) Potential molecular functions of ID family genes were identified using FunRich. (C) Potential protein domains of ID family genes were identified using FunRich.
Discussion
The development of molecular biology technology and bioinformatics has helped to understand the molecular biological characteristics of ID family members and their role in tumorigenesis. However, to the best of our knowledge, there are few studies on the role and significance of ID family members in lung cancer. Therefore, the present study investigated the expression patterns, prognostic values and potential functions of ID family members in lung cancer using bioinformatics methods. Thus, the present results may facilitate the development of future studies to identify potential therapeutic targets in lung cancer.
The present study analyzed the expression levels of ID family members in human tumors using the Oncomine database. The present results suggested that all ID family members had low expression levels in lung cancer. Furthermore, the experimental results based on the GEPIA database showed that the expression levels of ID family members were significantly decreased in lung cancer tissues, including lung adenocarcinoma and lung squamous cell carcinoma. In addition, it was also found that the expression level of ID4 was significantly different in various tumor stages. Therefore, the present results suggested that the expression levels of ID family members may be linked to the pathogenesis of lung cancer. In relation to this, Zhou et al. reported that the mRNA expression levels of ID1, ID3 and ID4 were significantly lower in breast cancer tissues compared with normal tissues [30]. However, the present results are inconsistent with other previous results [8–10], and these differences may be due to the small sample size and discrepancies in detection methods amongst the different studies.
In order to understand the relationship between ID family members and lung cancer, the present study performed a prognostic analysis using the Kaplan–Meier Plotter. It was demonstrated that increased ID2/3/4 mRNA expression levels were associated with improved OS, FP and PPS. However, decreased ID1 mRNA expression level was associated with improved OS, FP and PPS. Therefore, the present results suggested that low mRNA expression level of ID1 or high mRNA expression levels of ID2/3/4 predicted an improved survival in patients with lung cancer. In addition, the present study assessed the associations of the ID mRNA expression levels with distinct clinical parameters for OS, including histology, clinical stages, pathological grades, AJCC stages, sex and smoking status. For histology, a low mRNA expression level of ID1 was significantly associated with favorable OS in the following: Adenocarcinoma, squamous cell carcinoma, patients with stage 1 lung cancer, Grade II, AJCC stage N0, sex and smoking history. However, low mRNA expression levels of ID2 showed an unfavorable OS in the following: Adenocarcinoma, patients with stage 1 lung cancer, female patients and patients without a smoking history. Moreover, a low mRNA expression level of ID4 was found to be associated with unfavorable OS in the following categories: Adenocarcinoma, patients with stage 1 lung cancer, sex and patients without a smoking history. It was also demonstrated that ID3 mRNA expression level was significantly associated with unfavorable OS in AJCC stage T4, while ID3 showed an association with a favorable OS in female patients. Thus, the present results suggested that ID1/2/4 mRNA expression levels were correlated with pathological type, sex and smoking history in patients with lung cancer.
Currently, it is not fully understood whether IDs act as an oncogenes or a tumor suppressor genes in different tumor types due to a lack of identification of genetic alternations in ID genes. Li et al. found that 26.7% of ID3−/− mice developed lymphoma, while none of the ID3+/+ or ID3+/− mice had lymphoma. Therefore, a deficiency of the ID3 gene increases the possibility of γδ T-cell lymphoma [31]. However, to the best of our knowledge, there was no previous studies investigating genetic alterations of ID genes in other cancer types. Thus, the present study evaluated the mutations of ID family members in patients with lung cancer using the cBioPortal database. The present results suggested that there were mutations in ID1/2/3/4, which were predominately amplification and deep deletion mutations.
Lung cancer not only is caused by the differential expression of ID family members, but also may be caused by the interaction between related genes [32]. Therefore, the present study constructed PPI networks using STRING data for individual ID family members. Furthermore, the possible functions of ID related genes were investigated using Funrich software, including biological process, molecular function and protein domain. It was found that ID1/2/4 were significantly enriched in ‘Regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolis’, ‘Transcription factor activity’ and ‘HLH domain’. However, ID3 was not significantly enriched in the above factors. Therefore, the present results indicated that ID family members may be involved in cell metabolism and transcription regulation.
Transcription factors are a group of sequence-specific binding proteins that can activate or inhibit transcription via a transactivation or transrepression domains [33]. Previous studies have indicated that transcription factors are involved in regulating cell differentiation, proliferation and apoptosis, and play significant roles in the occurrence and development of tumors [34]. In the human genome, the C2H2zincfinger domain, homeodomain and HLH domain are the main types of transcription factor, and accounted for >80% of human genome [35]. Therefore, it is important to study the role of transcription factors in lung cancer. A nucleotide is an essential nutrient required to maintain the rapid proliferation of tumor cells, and nucleotide synthesis is regulated by many enzymes, genes and various metabolic pathways [36,37]. Moreover, previous studies have reported that inactivation of tumor suppressors and activation of oncogenes can promote the occurrence and development of tumors by regulating the biosynthesis of nucleotides [38]. Therefore, the mechanism of cell metabolism and transcriptional regulation of ID family genes, and related genes in the pathogenesis of lung cancer may be an important focus for future research. In addition, the present study supports the initiation of future studies to investigate the mechanism of tumor occurrence and development, to facilitate the development of prevention and treatment strategies.
In conclusion, the present study investigated the role of ID family members in lung cancer, in relation to mRNA expression levels, prognostic values, genetic mutations and functional enrichment analysis. The present results suggested that genetic mutations and mRNA expression levels were abnormal in patients with lung cancer. Furthermore, decreased ID1 or increased ID2/3/4 expression levels predicted an improved survival, and it was found that these proteins were involved in cell metabolism and transcription regulation. Therefore, ID family members may be used as biomarkers for the occurrence and prognosis of lung cancer. However, further study is required to assess the expression levels and molecular mechanisms of ID family members.
Abbreviations
- cBioPortal
cBio Cancer Genomics Portal
- FP
progression-free survival
- GEPIA
Gene Expression Profiling Interactive Analysis
- HLH
helix–loop–helix
- ID
inhibitor of differentiation/DNA-binding
- NSCLC
non-small cell lung cancer
- OS
overall survival
- PPI
protein–protein interaction
- PPS
post-progression survival
- SCLC
small cell lung cancer
- STRING
Search Tool for the retrieval of interacting Genes
Contributor Information
Chunfang Wang, Email: wangchunfang@sxmu.edu.cn.
Xueqing Wu, Email: xueqingwu416@126.com.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The present work was supported by National Key Research and Development Program [grant number 2018YFC1002103].
Author Contribution
Suming Xu, Chunfang Wang and Xueqing Wu conceived and designed the study. Yanhong Li and Lei Zhang prepared the figures and tables. Suming Xu and Yaoqin Wang analyzed and interpreted the data. Suming Xu and Yaoqin Wang drafted the manuscript. Chunfang Wang and Xueqing Wu revised the manuscript. All authors read and approved the final manuscript.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M. et al. (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Balata H., Fong K.M., Hendriks L.E., Lam S., Ostroff J.S., Peled N. et al. (2019) Prevention and Early Detection for NSCLC: Advances in Thoracic Oncology 2018. J. Thorac. Oncol. 14, 1513–1527 10.1016/j.jtho.2019.06.011 [DOI] [PubMed] [Google Scholar]
- 3.Torre L.A., Siegel R.L. and Jemal A. (2016) Lung Cancer Statistics. Adv. Exp. Med. Biol. 893, 1–19 10.1007/978-3-319-24223-1_1 [DOI] [PubMed] [Google Scholar]
- 4.Norton J.D. (2000) ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J. Cell Sci. 113, 3897–3905 [DOI] [PubMed] [Google Scholar]
- 5.Perk J., Iavarone A. and Benezra R. (2005) Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer 5, 603–614 10.1038/nrc1673 [DOI] [PubMed] [Google Scholar]
- 6.Lasorella A., Benezra R. and Iavarone A. (2014) The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat. Rev. Cancer 14, 77–91 10.1038/nrc3638 [DOI] [PubMed] [Google Scholar]
- 7.Roschger C. and Cabrele C. (2017) The Id-protein family in developmental and cancer-associated pathways. Cell Commun. Signal. 15, 7 10.1186/s12964-016-0161-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamalian L., Gosney J.R., Forootan S.S., Foster C.S., Bao Z.Z., Beesley C. et al. (2008) Increased expression of Id family proteins in small cell lung cancer and its prognostic significance. Clin. Cancer Res. 14, 2318–2325 10.1158/1078-0432.CCR-07-4716 [DOI] [PubMed] [Google Scholar]
- 9.Cheng Y.J., Tsai J.W., Hsieh K.C., Yang Y.C., Chen Y.J., Huang M.S. et al. (2011) Id1 promotes lung cancer cell proliferation and tumor growth through Akt-related pathway. Cancer Lett. 307, 191–199 10.1016/j.canlet.2011.04.003 [DOI] [PubMed] [Google Scholar]
- 10.Chen D., Forootan S.S., Gosney J.R., Forootan F.S. and Ke Y. (2014) Increased expression of Id1 and Id3 promotes tumorigenicity by enhancing angiogenesis and suppressing apoptosis in small cell lung cancer. Genes Cancer 5, 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., Li Y., Wang B., Ma Y. and Chen P. (2017) Id-1 promotes migration and invasion of non-small cell lung cancer cells through activating NF-kappaB signaling pathway. J. Biomed. Sci. 24, 95 10.1186/s12929-017-0400-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi K., Li Y., Li X., Lei X., Wang B., Zhang L. et al. (2016) Id4 promotes cisplatin resistance in lung cancer through the p38 MAPK pathway. Anticancer Drugs 27, 970–978 10.1097/CAD.0000000000000414 [DOI] [PubMed] [Google Scholar]
- 13.Rollin J., Blechet C., Regina S., Tenenhaus A., Guyetant S. and Gidrol X. (2009) The intracellular localization of ID2 expression has a predictive value in non small cell lung cancer. PLoS One 4, e4158 10.1371/journal.pone.0004158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B. et al. (2007) Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166–180 10.1593/neo.07112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z., Li C., Kang B., Gao G., Li C. and Zhang Z. (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy A., Lanczky A., Menyhart O. and Gyorffy B. (2018) Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 8, 9227 10.1038/s41598-018-27521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyorffy B., Surowiak P., Budczies J. and Lanczky A. (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8, e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A. et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. et al. (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A. et al. (2015) FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 15, 2597–2601 10.1002/pmic.201400515 [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharjee A., Richards W.G., Staunton J., Li C., Monti S., Vasa P. et al. (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. U.S.A. 98, 13790–13795 10.1073/pnas.191502998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beer D.G., Kardia S.L., Huang C.C., Giordano T.J., Levin A.M., Misek D.E. et al. (2002) Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 8, 816–824 10.1038/nm733 [DOI] [PubMed] [Google Scholar]
- 23.Su L.J., Chang C.W., Wu Y.C., Chen K.C., Lin C.J., Liang S.C. et al. (2007) Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics 8, 140 10.1186/1471-2164-8-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landi M.T., Dracheva T., Rotunno M., Figueroa J.D., Liu H., Dasgupta A. et al. (2008) Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One 3, e1651 10.1371/journal.pone.0001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selamat S.A., Chung B.S., Girard L., Zhang W., Zhang Y., Campan M. et al. (2012) Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 22, 1197–1211 10.1101/gr.132662.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okayama H., Kohno T., Ishii Y., Shimada Y., Shiraishi K., Iwakawa R. et al. (2012) Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 72, 100–111 10.1158/0008-5472.CAN-11-1403 [DOI] [PubMed] [Google Scholar]
- 27.Hou J., Aerts J., den Hamer B., van Ijcken W., den Bakker M., Riegman P. et al. (2010) Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 5, e10312 10.1371/journal.pone.0010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garber M.E., Troyanskaya O.G., Schluens K., Petersen S., Thaesler Z., Pacyna-Gengelbach M. et al. (2001) Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl. Acad. Sci. U.S.A. 98, 13784–13789 10.1073/pnas.241500798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stearman R.S., Dwyer-Nield L., Zerbe L., Blaine S.A., Chan Z., Bunn P.A. Jr et al. (2005) Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am. J. Pathol. 167, 1763–1775 10.1016/S0002-9440(10)61257-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X.L., Zeng D.E., Ye Y.H., Sun S.M., Lu X.F., Liang W.Q. et al. (2018) Prognostic values of the inhibitor of DNAbinding family members in breast cancer. Oncol. Rep. 40, 1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Maruyama T., Zhang P., Konkel J.E., Hoffman V., Zamarron B. et al. (2010) Mutation of inhibitory helix-loop-helix protein Id3 causes gammadelta T-cell lymphoma in mice. Blood 116, 5615–5621 10.1182/blood-2010-03-274506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim M.S., Park T.I., Lee Y.M., Jo Y.M. and Kim S. (2013) Expression of Id-1 and VEGF in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 6, 2102–2111 [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert M., Jambon S., Depauw S. and David-Cordonnier M.H. (2018) Targeting Transcription Factors for Cancer Treatment. Molecules 23, 1479 10.3390/molecules23061479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sever R. and Brugge J.S. (2015) Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 5, 10.1101/cshperspect.a006098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A. and Luscombe N.M. (2009) A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 10, 252–263 10.1038/nrg2538 [DOI] [PubMed] [Google Scholar]
- 36.DeBerardinis R.J., Lum J.J., Hatzivassiliou G. and Thompson C.B. (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 10.1016/j.cmet.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 37.Aird K.M., Zhang G., Li H., Tu Z., Bitler B.G., Garipov A. et al. (2013) Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 3, 1252–1265 10.1016/j.celrep.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villa E., Ali E.S., Sahu U. and Ben-Sahra I. (2019) Cancer Cells Tune the Signaling Pathways to Empower de Novo Synthesis of Nucleotides. Cancers (Basel) 11, 688 10.3390/cancers11050688 [DOI] [PMC free article] [PubMed] [Google Scholar]