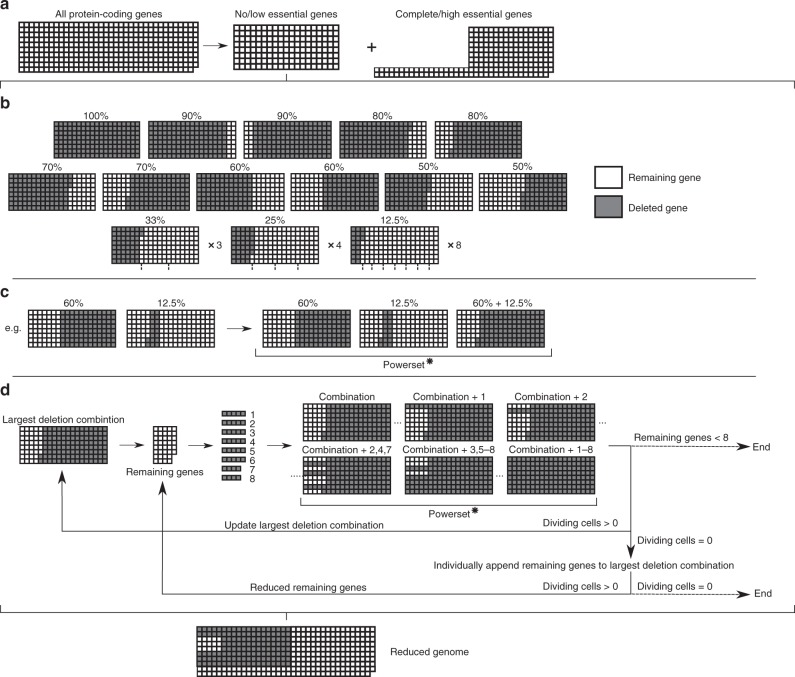
Fig. 1. Minesweeper algorithm for genome design.
a In silico single-gene knockouts are conducted to identify no/low essential genes (whose knockout does not prevent cell division). b 26 deletion segments, ranging in size from 100 to 12.5% of the no/low essential genes, are simulated. Grey indicates a gene deletion, white indicates a remaining gene. Deletion segments that on removal do not prevent division go to the next stage. c The largest deletion segment is matched with all division-producing, non-overlapping segments. A powerset (all possible unique combinations of this set of matched deletion segments) is generated and each combination simulated. Combination segments that do not prevent division go to the next stage. d The largest combination segment determines the remaining no/low essential genes that have not been deleted. These remaining genes are divided into eight groups (see Methods), a powerset generated for these eight groups, and each member of the powerset individually appended to the current largest deletion combination and simulated. If none of these simulations on removal produces a dividing cell, the remaining genes are appended as single knockouts to the current largest deletion combination, removed and simulated. The individual remaining genes that do not produce a dividing cell are temporarily excluded and a reduced remaining gene list produced. Details of simulations settings are available in the Methods section. Simulation data generated by the Minesweeper algorithm is available37 (see Data availability section). Powerset* = the complete powerset is not displayed here.