Figure 2.
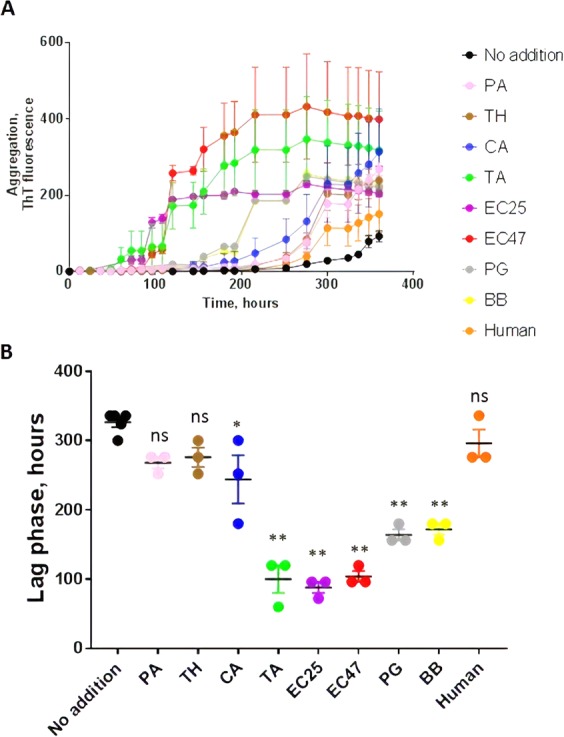
Effect of DNA extracted from diverse sources on tau aggregation. To study the effect of DNA on tau aggregation, monomeric tau (22 µM) under the conditions described in Fig. 1E, was incubated with preparations containing 100 ng of DNA extracted from different bacterial species including Pseudomonas aeruginosa (PA), Tetzosporium hominis (TH), Tetzerella alzheimeri (TA), Escherichia coli ATCC 25922 (EC25), Escherichia coli ATCC 472217 (EC47), Porphyromonas gingivalis (PG), Borrelia burgdorferi (BB). We also incubated tau with same amount of DNA extracted from Candida albicans (CA) and human samples. In all experiments the signal at time zero, corresponding to buffer + DNA + heparin + ThT + monomeric tau was substracted from the values. (A) tau aggregation was monitored over time by ThT fluorescence. Data corresponds to the average ± standard error of experiments done in triplicate (except for control without seeds that was performed in quintuplicate). (B) The lag phase, estimated as the time in which ThT fluorescence was higher than the threshold of 40 arbitrary units, was calculated for each experiment. The points represent the values obtained in each of the replicates. Data was analyzed by one-way ANOVA, followed by Tukey multiple comparison post-test. *P < 0.01; **P < 0.001.
