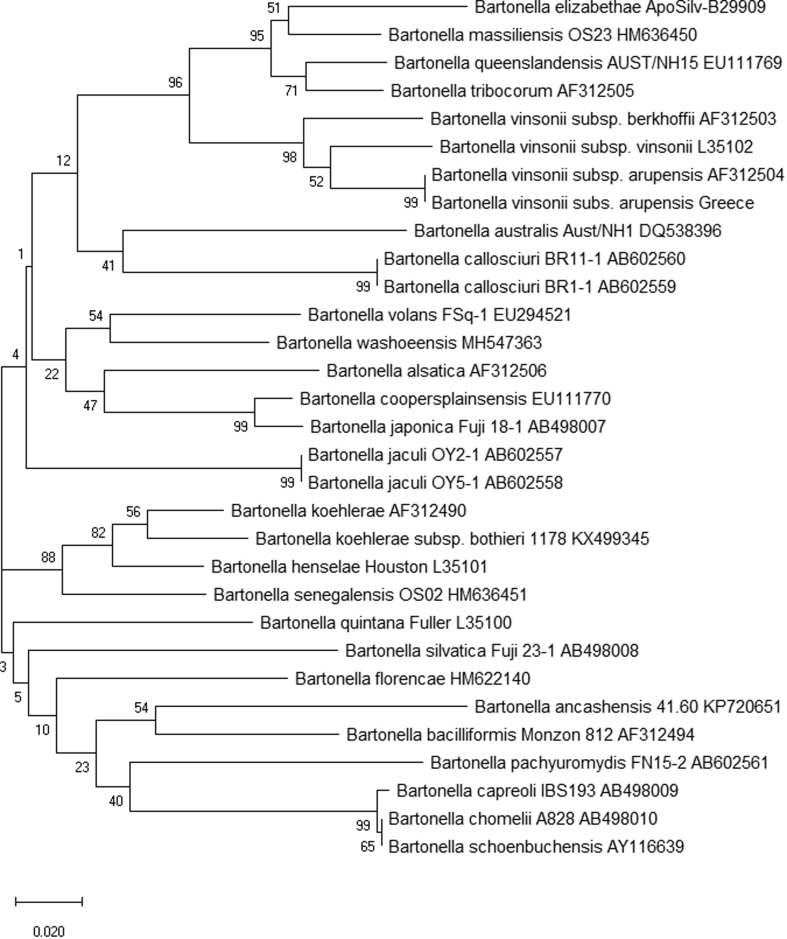
Fig. 1.
Internal transcribed spacer (ITS) phylogeny for a 408-bp fragment of the 16S–23S intergenic linker region of 33 Bartonella species. The evolutionary history was inferred using the neighbour-joining method. The optimal tree with the sum of branch length 1.67495836 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated (complete deletion option) [61].