Abstract
Fast response to environmental changes plays a key role in the transmission and pathogenesis of Yersinia enterocolitica. Osmoregulated periplasmic glucans (OPGs) are known to be involved in environmental perception of several Enterobacteriaceae pathogens; however, the biological function of OPGs in Y. enterocolitica is still unclear. In this study, we investigated the role of OPGs in Y. enterocolitica by deleting the opgGH operon encoding enzymes responsible for OPGs biosynthesis. Complete loss of OPGs in the ΔopgGH mutant resulted in decreased motility, c-di-GMP production, biofilm formation and smaller cell size, whereas the overproduction of OPGs through restoration of opgGH expression promoted c-di-GMP/biofilm production and increased antibiotic resistance of Y. enterocolitica. Gene expression analysis revealed that opgGH deletion reduced transcription of flhDC, ftsAZ, hmsT and hmsHFRS genes regulated by the Rcs phosphorelay system, whereas additional deletion of rcs family genes (rcsF, rcsC, or rcsB) reversed this effect and restored motility and c-di-GMP/biofilm production but further reduced cell size. Furthermore, disruption of the Rcs phosphorelay increased the motility and promoted the induction of biofilm and c-di-GMP production regulated by OPGs through upregulating the expression of flhDC, hmsHFRS, and hmsT. However, deletion of genes encoding the EnvZ/OmpR phosphorelay downregulated the flhDC, hmsHFRS and hmsT expression, leading to the decreased motility and prevented the induction of biofilm and c-di-GMP production regulated by OPGs. These results indicated that Rcs phosphorelay had the effect on OPGs-mediated functional responses in Y. enterocolitica. Our findings disclose part of the biological role of OPGs and the underlying molecular mechanisms associated with Rcs system in the regulation of the pathogenic phenotype in Y. enterocolitica.
Keywords: Yersinia enterocolitica, osmoregulated periplasmic glucans, Rcs phosphorelay, EnvZ/OmpR phosphorelay, gene expression, pathogenic phenotype
Introduction
Yersinia enterocolitica is a common foodborne pathogen which causes a zoonotic disease called yersiniosis manifested in humans by acute gastroenteritis and sometimes more serious conditions such as pseudoappendicitis and even sepsis (Huovinen et al., 2010; Bari et al., 2011; Fabrega and Vila, 2012). In the reports of the European Food Safety Authority (EFSA), Y. enterocolitica is listed as the third most common enteropathogen after Campylobacter and Salmonella (Zadernowska et al., 2014). Y. enterocolitica is widely distributed in the environment and can be found in soil, water, animals and various food products (Bari et al., 2011; Rahman et al., 2011), where constant changes in physicochemical conditions, including osmolarity, pH, temperature, light intensity, medium viscosity, and nutrient availability often threaten bacterial survival (Bottone, 1997; Brzostek et al., 2012). These challenges are met by rapid adaptation of the pathogen to varying growth conditions provided by two-component regulatory systems (TCSs), also called phosphorelays, as they sense the extracellular signals and perform cascade phosphorylation in response, thus regulating the expression of genes related to flagellar synthesis, biofilm formation, and virulence (Clarke, 2010; Clarke and Voigt, 2011; Brzostek et al., 2012; Liu et al., 2017).
EnvZ/OmpR is a classical TCS present in many bacteria, including Y. enterocolitica (Brzostek et al., 2012). Under environmental stimuli, transmembrane histidine kinase EnvZ (sensor) is autophosphorylated and in turn phosphorylates a transcription factor OmpR, which regulates the expression of genes controlling numerous bacterial cell functions, including outer membrane permeability (Russo and Silhavy, 1991), flagella synthesis (Raczkowska et al., 2011), biofilm formation (Pruss, 2017), and pH tolerance (Bang et al., 2000). It has been reported that in Y. enterocolitica, OmpR could play an important role in controlling the virulence properties (Dorrell et al., 1998; Brzostek et al., 2003), act as the response regulator for osmolarity-regulated porins and Yop proteins (Brzostek et al., 2003), negatively regulate invasion gene expression (Brzostek et al., 2007) and positively control motility and flhDC expression (Raczkowska et al., 2011). The EnvZ/OmpR phosphorelay has been reported to be a central regulation system of several cellular responses in Y. enterocolitica (Brzostek et al., 2012).
The regulator of capsule synthesis (Rcs) phosphorelay is an atypical TCS restricted to Enterobacteria (Wall et al., 2018). This system is composed of three core proteins, the transmembrane sensor kinase RcsC, transmembrane protein RcsD, and response regulator RcsB, and could be activated by an outer membrane-associated lipoprotein RcsF in response to stress (Castanie-Cornet et al., 2006). As a result of this phosphorelay, RcsB is phosphorylated and then interacts with a conserved motif in target genes, thus regulating their transcription (Clarke, 2010). In several Enterobacteria spp., the Rcs system downregulates the flhDC master operon encoding activators required for expression of the flagellar apparatus genes and upregulates the ftsAZ operon needed for cell division (Bontemps-Gallo et al., 2013). A recent study reported that in Yersinia pestis, the Rcs system inhibited biosynthesis of cyclic dimeric guanosine monophosphate (c-di-GMP) and biofilm formation by directly repressing transcription of the hmsT gene and the hmsHFRS operon (Fang et al., 2015). The Rcs system has also been reported in Y. enterocolitica; however, the signaling mechanism underlying its participation in environmental responses is still unclear.
Osmoregulated periplasmic glucans (OPGs) formerly known as membrane-derived oligosaccharides are polymers of D-glucose connected through β-linkage (Bontemps-Gallo and Lacroix, 2015), which are present in the cell envelope of Gram-negative bacteria (Bontemps-Gallo et al., 2017). As the name indicates, OPGs are osmoregulated and their synthesis and accumulation decreases with the increase in osmolarity (Bhagwat et al., 2009; Bontemps-Gallo et al., 2013). The synthesis of OPGs requires the opgGH operon in Enterobacteria or its functional homologs ndvAB/chvAB/cgs in other bacterial species (Bontemps-Gallo et al., 2017). Deletion of these operons causes a total loss of OPGs, resulting in a pleiotropic phenotype (Bontemps-Gallo and Lacroix, 2015). OPGs represent virulence factors of many pathogenic bacteria; thus, Dickeya dadantii and Salmonella lacking OPGs exhibited a non-virulent phenotype (Page et al., 2001; Bhagwat et al., 2009). In Escherichia coli, OPGs were shown to regulate chemotaxis, motility and intercellular signaling (Weissborn et al., 1992), whereas in Brucella abortus, OPGs secretion was required in the early stage of cell infection and invasion (Briones et al., 2001), and in Pseudomonas aeruginosa, the presence of OPGs facilitated biofilm formation and conferred higher antibiotic resistance (Mah et al., 2003). Furthermore, it has been reported that mutations in the operon responsible for structural assembly of OPGs led to activation of the Rcs phosphorelay. Thus, in E. coli, the lack of OPGs was reported to trigger the Rcs phosphorelay, which resulted in suppression of bacterial motility (Girgis et al., 2007), whereas OPGs-negative D. dadantii totally lost virulence and motility, which was due to constitutive activation of the Rcs phosphorelay (Bouchart et al., 2010; Bontemps-Gallo et al., 2013; Madec et al., 2014). These data indicate that OPGs play an important role in the pleiotropic phenotype of pathogenic bacteria, showing a regulatory effect on TCSs.
As a member of the Enterobacteriaceae family, Y. enterocolitica also contains the complete opgGH gene cluster as revealed by genome-wide sequencing (Thomson et al., 2006), but the functional significance of opgGH in Y. enterocolitica is largely unknown; the only evidence of it is that OpgH is required in the early stage of infection (Young and Miller, 1997). Except for Y. enterocolitica, there are other two human pathogenic species in the genus Yersinia, Yersinia pseudotuberculosis, and Y. pestis (Kim et al., 2008). Y. pseudotuberculosis causes gastroenteritis, while Y. pestis is the causative agent of plague. Recent studies found that Y. pseudotuberculosis 2777 (serotype O:1) and 2515 (serotype O:2) did not produce OPGs. With the deletion of opgGH, Y. pseudotuberculosis showed normal motility, biofilm formation and virulence, but smaller cell size (Quintard et al., 2015). Y. pestis lost the opgGH operon during its emergence from Y. pseudotuberculosis. After re-introducing opgGH into Y. pestis, no effect on flea proventricular blockage rate was found (Quintard et al., 2015). All these studies on the biological functions of opgGH in Enterobacteria spp. led us to explore the role of opgGH and its products in Y. enterocolitica.
In this study, we showed that OPGs were present in Y. enterocolitica (biotype 1B and serotype O:8) and that deletion of the opgGH operon generated a pleiotropic phenotype. Gene expression analysis indicated that OPGs deficiency was correlated with the activation of the Rcs phosphorelay, whereas OPGs overexpression enhanced c-di-GMP production and biofilm formation and conferred higher antibiotic resistance to Y. enterocolitica. It was also found that Rcs and EnvZ/OmpR phosphorelays had opposite effects on the regulation of OPGs-induced c-di-GMP production and biofilm formation in Y. enterocolitica. These findings disclose part of the biological role of OPGs and further understanding of Rcs and EnvZ/OmpR phosphorelays in the regulation of the pathogenic phenotype in Y. enterocolitica.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH5α used as the host bacteria in plasmid construction was cultured at 37°C in lysogeny broth (LB) consisting of 5 g/L yeast extract, 10 g/L tryptone, and 5 g/L NaCl. Y. enterocolitica ATCC23715 (biotype 1B and serotype O:8) was used as the parent strain for construction of Y. enterocolitica mutants; the bacteria were cultured at 26°C in different growth media: LB, LBNS (LB without salts) and LNNS (LBNS broth diluted 1:8 in distilled water). Ampicillin (100 μg/ml), chloramphenicol (16 μg/ml), cefsulodin (15 μg/ml), irgasan (4 μg/ml), and novobiocin (2.5 μg/ml) were added as required.
TABLE 1.
Strains and plasmids used in this study.
| Strains and plasmids | Relevant characteristics | Sources |
| Y. enterocolitica | ||
| ATCC23715 | WT, serotype O:8, Biotype 1B, pYV– | Lab stock |
| YE-1 | △opgGH | This study |
| YE-2 | △opgGH, PBADopgGH; Ampr | This study |
| YE-B | △rcsB | Meng et al., 2019 |
| YE-C | △rcsC | This study |
| YE-F | △rcsF | This study |
| YE-1B | △opgGH, △rcsB | This study |
| YE-1C | △opgGH, △rcsC | This study |
| YE-1F | △opgGH, △rcsF | This study |
| YE-2B | △opgGH, △rcsB, PBADopgGH; Ampr | This study |
| YE-2C | △opgGH, △rcsC, PBADopgGH; Ampr | This study |
| YE-2F | △opgGH, △rcsF, PBADopgGH; Ampr | This study |
| YE-Z | △envZ | This study |
| YE-R | △ompR | Meng et al., 2019 |
| YE-1Z | △opgGH, △envZ | This study |
| YE-1R | △opgGH, △ompR | This study |
| YE-2Z | △opgGH, △envZ, PBADopgGH; Ampr | This study |
| YE-2R | △opgGH, △ompR, PBADopgGH; Ampr | This study |
| E. coli | ||
| S17-1 λpir | recA1, thi, pro, hsdR-M +, RP4:2-Tc:Mu–Kan:Tn7, λpir | Lab stock |
| DH5a | F–, φ80lacZ△M15, △(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17 (rk–,mk+), phoA, supE44, λ–, thi-1, gyrA96, relA1 | Lab stock |
| Plasmids | ||
| pDS132 | Conditional replication vector; | Lab stock |
| R6K origin, mobRK4 transfer origin, sucrose-inducible-sacB; Cmr | ||
| pDS132-△opgGH | Upstream and downstream opgGH fragments | This study |
| were cloned into pDS132; Cmr | ||
| pDS132-△rcsB | Upstream and downstream rcsB fragments | This study |
| were cloned into pDS132; Cmr | ||
| pDS132-△rcsC | Upstream and downstream rcsC fragments | This study |
| were cloned into pDS132; Cmr | ||
| pDS132-△rcsF | Upstream and downstream rcsF fragments | This study |
| were cloned into pDS132; Cmr | ||
| pDS132-△envZ | Upstream and downstream envZ fragments | This study |
| were cloned into pDS132; Cmr | ||
| pDS132-△ompR | Upstream and downstream ompR fragments | This study |
| were cloned into pDS132; Cmr | ||
| pBAD24 | AraC, promoter PBAD; Ampr | Lab stock |
| pBAD24-opgGH | AraC, PBAD opgGH; Ampr | This study |
Amp, ampicillin; Cm, chloramphenicol; r, resistance.
Plasmid Construction
To construct pDS132-ΔopgGH, fragments upstream and downstream of the opgGH gene were amplified from the Y. enterocolitica genome using primers opgGH-up-F/opgGH-up-R and opgGH-down-F/opgGH-down-R. The upstream and downstream fragments were fused and amplified by fusion PCR with primers opgGH-up-F/opgGH-down-R; the resultant long fragment was digested with SphI and SacI and ligated into pDS132 digested with the same enzymes to yield pDS132- ΔopgGH. The same approach was also used to construct pDS132-ΔrcsF, pDS132-ΔrcsC, pDS132-ΔrcsB, pDS132-ΔenvZ, and pDS132-ΔompR with the corresponding primers.
To construct pBAD24-opgGH, the opgGH fragment was amplified from the Y. enterocolitica genome using primers p-opgGH-F/p-opgGH-R, digested with SalI and HindIII, and inserted into pBAD24 digested with the same enzymes. The resultant pBAD24-opgGH plasmid contained the opgGH operon controlled by the araBAD promoter (PBADopgGH).
Strain Construction
For construction of the opgGH knockout strain, the suicide plasmid pDS132-ΔopgGH was introduced into E. coli S17-1λpir by electroporation and then mobilized into Y. enterocolitica by conjugation. The strategy used for gene deletion in the Y. enterocolitica chromosome was based on the two-step homologous recombination procedure described previously (Schafer et al., 1994); the process of genetic manipulation is shown in Supplementary Figure 1. The same approach was used for deletion of the rcsF, rcsC, rcsB, envZ, and ompR genes in the ΔopgGH mutant and wild-type strain, respectively. The mutants were verified by polymerase chain reaction (PCR) and further confirmed by gene sequencing. The pBAD24-opgGH plasmid was used to transform ΔopgGH, ΔopgGH-ΔrcsF, ΔopgGH-ΔrcsC, ΔopgGH-ΔrcsB,ΔopgGH-ΔenvZ, and ΔopgGH-ΔompR mutant strains by electroporation to yield ΔopgGH-PBADopg GH, ΔopgGH-ΔrcsF-PBADopgGH, ΔopgGH-ΔrcsC-PBADopgGH, ΔopgGH-ΔrcsB-PBADopgGH,ΔopgGH-ΔenvZ-PBADopgGH, and ΔopgGH-ΔompR-PBADopgGH strains. All primers used for strain and plasmid construction are listed in Supplementary Table 1.
Large-Scale Purification and Determination of OPGs
Osmoregulated periplasmic glucans extraction, purification, and determination were based on the method as described previously (Bontemps-Gallo et al., 2013). To maximize the yield of OPGs, bacteria were grown in 8 L of LNNS medium supplemented with 0.6 g/L L-arabinose until the exponential phase, collected by centrifugation at 8,000 × g for 15 min at 4°C, resuspended in 200 ml of distilled water, and treated with 5% trichloroacetic acid. After centrifugation at 8,000 × g for 15 min at 4°C, the supernatant was collected and OPGs were extracted by charcoal adsorption, eluted with 15% aqueous pyridine, concentrated by rotary evaporation to a volume of 2 ml, and fractionated by gel filtration on a Bio-Gel P-4 column (1.6 cm × 55 cm; Bio-Rad). OPGs were eluted with 0.5% acetic acid at a flow rate of 15 ml/h, and 1.5-ml fractions were collected. Fractions containing OPGs were pooled and OPGs content was determined by a colorimetric method using the anthrone reagent (Spiro, 1966). Whole-cell protein amount was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific). Final OPGs content was expressed as μg/mg of protein.
Growth Assay
Growth rates of wild-type, ΔopgGH mutant and ΔopgGH-PBADopgGH strains were determined in LB, LBNS, and LNNS media. Single colonies were inoculated into appropriate medium and incubated overnight at 26°C with shaking. The starter cultures were diluted in 100 ml medium to OD600 about 0.05 and incubated in 250-ml flasks in a rotatory shaker at 180 rpm. Cell growth was measured at 600 nm in a spectrophotometer (Puxi Universal, Co., Ltd., Beijing).
Motility Assay
Swim motility experiments were performed in LNNS plates containing 0.35% agar. Bacteria were grown overnight in LB at 26°C, diluted to OD600 of 1. And 1 μl of diluted seed cultures were inoculated into swim agar plates and incubated at 26°C. Bacterial swim diameters were measured after 48-h incubation.
Biofilm Assay
Overnight cultures of Y. enterocolitica strains were inoculated into a 96-well plate containing 200 μl LNNS medium per well (initial OD600 about 0.05) and incubated at 26°C; six replicates per condition were used. Growth medium was renewed every 24 h. After 24, 48 and 72 h of incubation, growth medium was removed, and the formed biofilms were washed twice with PBS and stained with crystal violet staining solution (0.1%). Wells were washed with water and treated with ethanol-acetone solution to release the dye absorbed in the biofilm, which was then measured at 595 nm.
Measurement of Bacterial Cell Size
Bacteria grown to the mid-log phase in LNNS medium were immobilized on 200-mesh copper grids, fixed in 0.5% glutaraldehyde for 5 min, washed with water and air-dried. And the cell length was visualized and measured using a JEM-1230 transmission electron microscopy. Bacterial cells were magnified 10,000 times with approximately 30–50 cells per field of view. The cell length of each strain was the average of 100 measurements performed using iTEM software.
Extraction and Quantification of c-di-GMP
Bacteria were grown to the exponential phase in LNNS medium and 2 ml of planktonic culture with OD600 about 0.5 was centrifuged at 10,000 × g for 10 min at 4°C. The cell pellet was washed twice with ice-cold PBS, resuspended in 2 ml ice-cold PBS, incubated at 100°C for 5 min, and sonicated for 15 min (power 100%, frequency 37 kHz) in an ice-water bath. After centrifugation, the supernatant containing extracted c-di-GMP was collected and the pellet was resuspended in 2 ml ice-cold PBS; the extraction procedure described above was repeated twice. The extracts were concentrated by cooling evaporation at 4°C to a volume of 500 μl and intracellular c-di-GMP levels were determined by the c-di-GMP enzyme-linked immunosorbent assay (ELISA) Kit (Mskbio, Beijing, China). Cell protein was determined by the BCA assay and c-di-GMP concentrations were expressed as pmol/mg of protein.
Antibiotic Susceptibility Assay
The sensitivity of Y. enterocolitica strains to chloramphenicol and polymyxin B was determined by the agar disk diffusion method as described by Clinical and Laboratory Standards Institute (CLSI, 2012) with some modifications. Briefly, indicated strains were grown to 0.5 McFarland standard and 200 μl of bacterial suspension was streaked over LNNS agar using sterile cotton swabs. Paper disks (6 mm in diameter) containing different concentrations of chloramphenicol or polymyxin B were placed in the center of the plate and the diameter of the clear zone around the disk was measured after 24-h incubation at 26°C.
RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
Total RNA was extracted from Y. enterocolitica strains grown to the exponential phase in LNNS medium using the TransZol Up Plus RNA Kit (TransGen, Beijing, China). The extracted RNA was then tested for its concentration and quality using a Nanodrop 2000c (Thermo). And cDNA was synthesized from 500 ng RNA using Quant Reverse Transcriptase in the presence of random primers (TransGen, Beijing, China). RT-qPCR was carried out using Real Master Mix (SYBR Green) and specific primers (Supplementary Table 2) in a Light Cycler 480 II (Roche, Basel, Switzerland) under the following cycling conditions: 5 min at 50°C, 30 s at 94°C, and 45 cycles of 5 s at 94°C, and 30 s at 60°C. Reactions were performed in triplicate. Relative transcription of the target genes was analyzed by the 2–Δ Δ Ct method described previously (Meng et al., 2016); the 16S rRNA gene was used as reference for normalization.
Statistical Analysis
One-way analysis of variance was performed in SPSS for Windows 20.0 (SPSS, Inc., Chicago, IL, United States).
Results
OPGs Were Required for Y. enterocolitica Growth in Low-Nutrient Low-Salt Medium
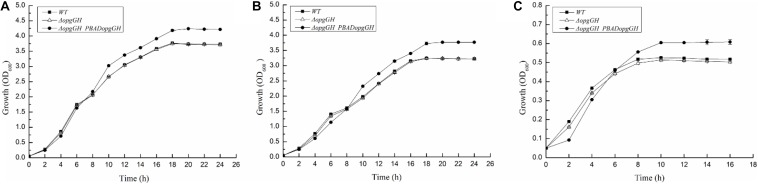
It has been reported that OPGs take part in environmental perception, and their abundance in the periplasm increases as the osmolarity of the medium decreases (Bhagwat et al., 2009; Bontemps-Gallo et al., 2013). Lack of OPGs has been confirmed to impair the optimal growth potential of Salmonella enterica serovar Typhimurium under low osmolarity conditions (Bhagwat et al., 2009). To determine the biological function of OPGs in Y. enterocolitica, we constructed an OPGs-deficient strain by deleting the opgGH operon responsible for OPGs synthesis from the Y. enterocolitica chromosome, and an OPGs-overproducing strain by transforming the ΔopgGH mutant with the pBAD24-opgGH plasmid containing the opgGH operon under the control of the inducible PBAD promoter. Expression levels of the opgG and opgH genes were upregulated by 605- and 474-fold, respectively, after addition of 0.6 g/L L-arabinose, indicating that the opgGH operon was upregulated by L-arabinose. To assess the effect of osmotic stress and nutrient deficiency on Y. enterocolitica growth, we measured growth characteristics of wild-type, ΔopgGH and ΔopgGH-PBADopgGH strains in LB, LBNS (LB, no salts) and LNNS (low nutrient, no salts) media. All tested strains were supplemented with 0.6 g/L L-arabinose. The deletion of opgGH did not affect bacterial growth in LB and LBNS, but caused a slight lag throughout the growth process in LNNS medium (P < 0.05), whereas opgGH overexpression led to cells reaching a significantly higher density in stationary phase in LB, LBNS, and LNNS media compared to wild-type strain (P < 0.01) (Figures 1A–C).
FIGURE 1.
Growth characteristics of the wild-type, ΔopgGH and ΔopgGH-PBADopgGH strains of Yersinia enterocolitica. Bacteria were grown in LB (A), LBNS (B), and LNNS (C) media supplemented with 0.6 g/L L-arabinose. The data are presented as the mean ± SD of at least three independent experiments.
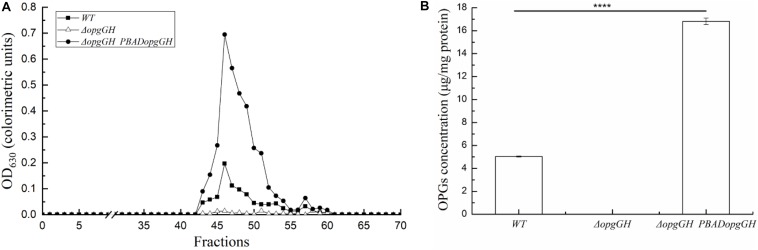
Deletion of opgGH showed impaired cell growth in LNNS medium, indicating that the growth of wild-type Y. enterocolitica was relatively dependent of OPGs synthesis under low-nutrient low-salt conditions. Then, we performed extraction and separation of OPGs from Y. enterocolitica cultured in LNNS medium. OPGs from wild-type and ΔopgGH-PBADopgGH strains were eluted from the BioGel P4 gel filtration column as a single major peak (fractions 43–55), whereas the elution profile of the ΔopgGH mutant lacked the corresponding peak (Figure 2A), suggesting the absence of OPGs in the ΔopgGH strain. Concentrations of OPGs in wild-type and ΔopgGH-PBADopgGH strains were 5.03 and 16.81 μg/mg protein, respectively (Figure 2B). This finding, in combination with growth characteristics, indicates that OPGs synthesized by opgGH-encoded enzymes regulate the growth of Y. enterocolitica under low-nutrient low-salt condition.
FIGURE 2.
Purification and quantification of OPGs from Y. enterocolitica. OPGs were extracted from wild-type, ΔopgGH and ΔopgGH-PBADopgGH strains grown to the exponential phase in LNNS medium supplemented with 0.6 g/L L-arabinose. (A) Gel filtration chromatography of OPGs. (B) Quantification of purified OPGs. The data are presented as the mean ± SD of at least three independent experiments. An asterisk indicates a significant difference with ****P < 0.0001.
OpgGH Deletion Decreased Y. enterocolitica Motility, c-di-GMP Production, Biofilm Formation, and Cell Size
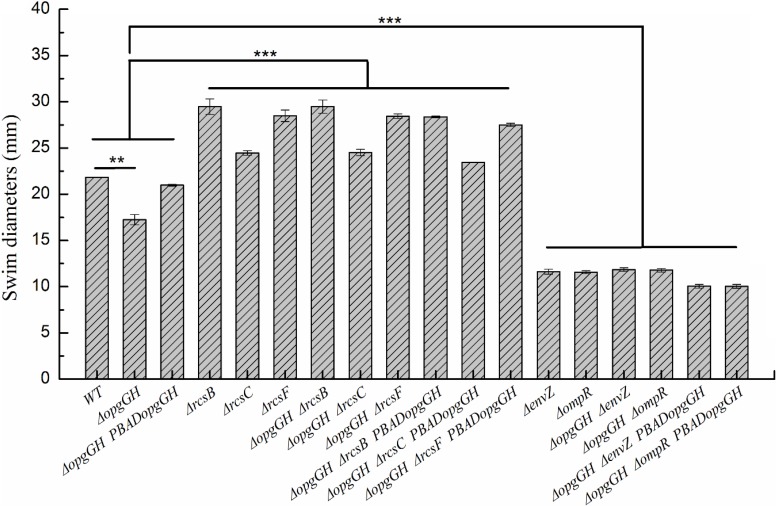
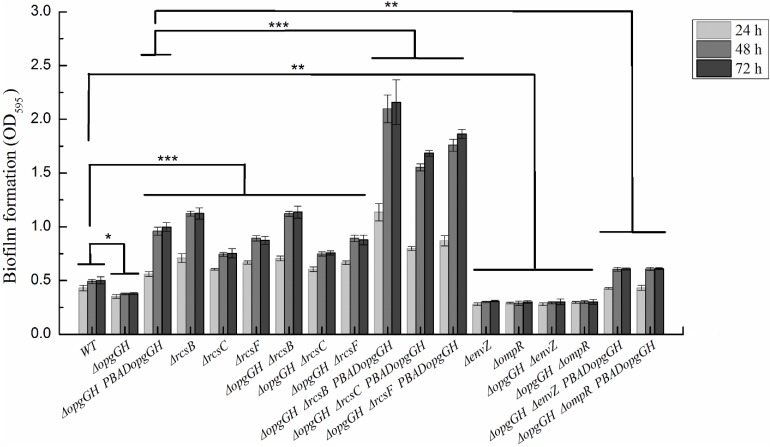
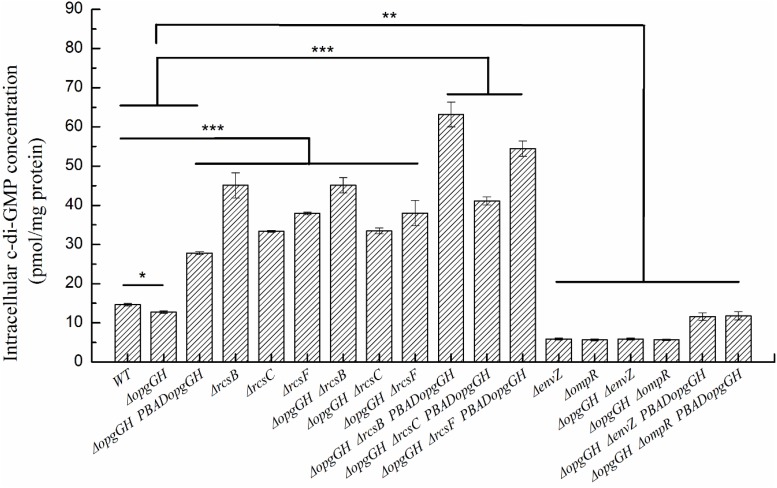
Previous evidence suggests that the lack of OPGs in Proteobacteria caused abnormal phenotypic changes, including decreased envelope stability, flagellar synthesis, biofilm formation, environmental tolerance, virulence, and pathogenicity (Bontemps-Gallo and Lacroix, 2015). Therefore, the effect of loss and overexpression of OPGs on the Y. enterocolitica phenotype was analyzed in LNNS medium. The results indicated that the ΔopgGH mutant decreased the swim diameter by 21% in LNNS agar, but motility was restored when the mutant was complemented with the wild-type opgGH operon (Figure 3 and Supplementary Figure 2). Furthermore, OPGs depletion reduced by 24% in biofilm formation, whereas OPGs overproduction almost doubled the yield of biofilms after 72 h of incubation as evidenced by crystal violet staining when compared to the wild-type strain (Figure 4 and Supplementary Figure 3). Similarly, the absence of OPGs resulted in 13% reduction, while OPGs overproduction led to a 90% increase in c-di-GMP production when compared to the wild-type strain (Figure 5).
FIGURE 3.
Changes in swim motility of wild-type and mutant strains in LNNS medium. Y. enterocolitica was grown in LNNS semisolid plates supplemented with 0.6 g/L L-arabinose at 26°C. Swim diameters were measured after 48 h of incubation. The data are presented as the mean ± SD of at least three independent experiments. An asterisk indicates a significant difference with ***P < 0.001, **P < 0.01.
FIGURE 4.
Biofilm formation in wild-type and mutant strains of Y. enterocolitica. Bacteria were cultured in LNNS medium supplemented with 0.6 g/L L-arabinose at 26°C. Biofilm formation was analyzed after 24, 48, and 72 h of incubation by staining with crystal violet and measuring absorbance at 595 nm. The data are presented as the mean ± SD of at least three independent experiments. An asterisk indicates a significant difference with ***P < 0.001, **P < 0.01, *P < 0.05.
FIGURE 5.
The intracellular c-di-GMP concentration in wild-type and mutant strains of Y. enterocolitica. Bacteria were cultured in LNNS medium supplemented with 0.6 g/L L-arabinose at 26°C. Intracellular c-di-GMP levels were analyzed in bacteria grown to the exponential phase. The data are presented as the mean ± SD of at least three independent experiments. An asterisk indicates a significant difference with ***P < 0.001, **P < 0.01, *P < 0.05.
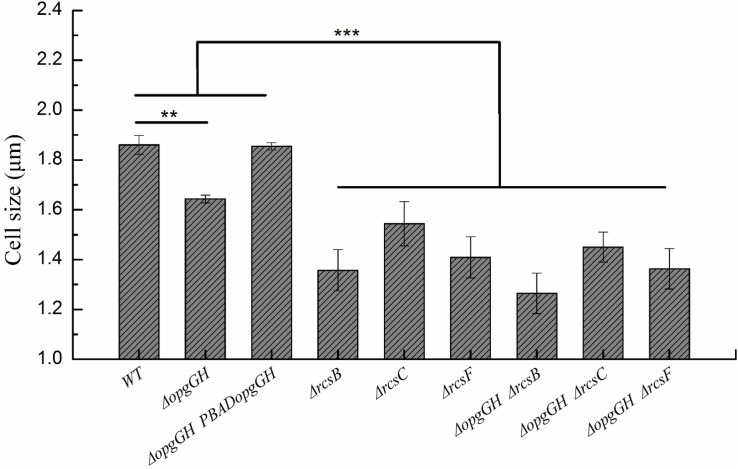
In addition to its ability to synthesize OPGs, the opgGH operon (opgH but not opgG) is also involved in the regulation of bacterial cell size through sequestration of FtsZ, a highly conserved tubulin-like cell division protein (Hill et al., 2013; Quintard et al., 2015). Therefore, we investigated the effect of opgGH loss on Y. enterocolitica cell shape during growth in LNNS medium. As shown in Figure 6 and Supplementary Figure 4, opgGH-deficient cells were about 12% smaller than wild-type cells, but the normal cell size was restored when mutant cells were complemented with opgGH, suggesting that opgGH (consider as a whole) controls cell size in Y. enterocolitica.
FIGURE 6.
Changes in the cell size of wild-type and mutant strains. Bacteria were grown to the mid-log phase in LNNS medium supplemented with 0.6 g/L L-arabinose and cell size was measured using transmission electron microscopy. The data are presented as the mean ± SD of at least three independent experiments. An asterisk indicates a significant difference with ***P < 0.001, **P < 0.01.
OPGs Overproduction Conferred Higher Antibiotic Resistance to Y. enterocolitica
Previous reports suggested that in addition to the presence in the periplasmic space, OPGs can be secreted from the cell and directly interact with antibiotics to protect bacteria and promote their survival (Mah et al., 2003; Bontemps-Gallo and Lacroix, 2015). To investigate the effect of OPGs on antibiotic sensitivity of Y. enterocolitica, we assessed the clearing zones produced by chloramphenicol, which inhibits protein synthesis, and polymyxin B, which destabilizes bacterial cell membrane. Visible from the results that Y. enterocolitica was more sensitive to chloramphenicol than to polymyxin B (Table 2). For chloramphenicol, both wild-type and ΔopgGH mutant strains appeared the clearing areas at a concentration of 0.125 mg/ml, whereas ΔopgGH-PBADopgGH strain were at 0.25 mg/ml. However, 0.25 mg/ml of polymyxin B resulted in the appearance of clearing areas of both wild-type and OPGs-deficient strains, whereas a higher concentration, at 0.5 mg/ml, was required for the emergence of inhibition zones in ΔopgGH-PBADopgGH strain. Further, zone of growth inhibition around a disk was also used to assess the sensitivity to antibiotics. The average sizes of clearing zones at different concentrations of the two antibiotics were slightly larger for the ΔopgGH mutant, but were markedly reduced after OPGs overproduction compared to the wild-type strain (P < 0.01) (Table 2), suggesting that OPGs increase resistance of Y. enterocolitica to at least two different classes of antibiotics.
TABLE 2.
Susceptibility of Y. enterocolitica strains to chloramphenicol and polymyxin B.
| Antibiotics | Strains | Average clearing zone (mm) | |||||
| 0.0625 mg/ml | 0.125 mg/ml | 0.25 mg/ml | 0.5 mg/ml | 1 mg/ml | 2 mg/ml | ||
| Chloramphenicol | WT | − | 6.88 ± 0.16 | 10.20 ± 0.12 | 13.52 ± 0.10 | 19.76 ± 0.11 | 21.48 ± 0.15 |
| △opgGH | − | 7.40 ± 0.27 | 11.04 ± 0.07 | 14.18 ± 0.29 | 20.28 ± 0.11 | 21.81 ± 0.17 | |
| △opgGH- PBADopgGH | − | − | 7.55 ± 0.31 | 10.45 ± 0.31 | 17.54 ± 0.38 | 20.47 ± 0.35 | |
| Polymyxin B | WT | − | − | 1.69 ± 0.10 | 2.93 ± 0.06 | 4.68 ± 0.24 | 5.14 ± 0.17 |
| △opgGH | − | − | 2.07 ± 0.04 | 3.34 ± 0.06 | 5.00 ± 0.11 | 5.45 ± 0.10 | |
| △opgGH- PBADopgGH | − | − | − | 1.50 ± 0.09 | 2.71 ± 0.06 | 3.78 ± 0.05 | |
The data are presented as the mean ± SD of at least three separate experiments. “−”, not detected.
Disruption of the Rcs Phosphorelay in the Y. enterocolitica Δ opgGH Mutant Restored Motility, c-di-GMP Production, and Biofilm Formation but Further Reduced Cell Size
It has been reported that activation of the Rcs phosphorelay triggered by OPGs deficiency suppressed motility of E. coli (Girgis et al., 2007) and attenuated virulence of D. dadantii (Bouchart et al., 2010), but the effects were reversed after deletion of any of the Rcs phosphorelay genes. In this study, we constructed ΔopgGH-ΔrcsF, ΔopgGH-ΔrcsC, and ΔopgGH-ΔrcsB double mutants to determine whether disruption of Rcs phosphorelay could reverse the effects caused by OPGs loss in Y. enterocolitica. As a result, the double mutants not only restored but showed even better swim motility than the wild-type strain. The ΔopgGH-ΔrcsF, ΔopgGH-ΔrcsC, and ΔopgGH-ΔrcsB double mutants had increased swim diameter by 30, 12, and 35%, respectively, compared to the wild-type strain (Figure 3 and Supplementary Figure 2). In addition, the swim diameter of ΔrcsF, ΔrcsC, and ΔrcsB single mutants were also performed to distinguish the effects of OPGs and Rcs phosphorelay on the motility of Y. enterocolitica. Deletion of rcsF, rcsC, and rcsB also increased swim diameter by 31, 12, and 35%, respectively, compared to the wild-type strain (Figure 3 and Supplementary Figure 2). However, there was no significant change in swim diameter between ΔopgGH-ΔrcsF/ΔopgGH-ΔrcsC/ΔopgGH-ΔrcsB double mutant and ΔrcsF/ΔrcsC/ΔrcsB single mutant (P > 0.05) (Figure 3), indicating that loss of OPGs will not affect the swim motility in the absence of Rcs phosphorelay in Y. enterocolitica.
Similarly, ΔopgGH-ΔrcsF, ΔopgGH-ΔrcsC, and ΔopgGH-ΔrcsB double mutants not only restored but further promoted biofilm and c-di-GMP production compared to the wild-type strain. The biofilm formation in ΔopgGH-ΔrcsF, ΔopgGH-ΔrcsC, and ΔopgGH-ΔrcsB double mutants were 76, 52% and 1.28-fold increase, respectively, compared to the wild-type strain after 72 h of incubation (Figure 4 and Supplementary Figure 3). The intracellular c-di-GMP production in ΔopgGH-ΔrcsF, ΔopgGH-ΔrcsC, and ΔopgGH-ΔrcsB double mutants were 1.59-, 1.28-, and 2.07-fold increase in comparison to the wild-type strain (Figure 5). Although ΔrcsF, ΔrcsC, and ΔrcsB single mutants upregulated the biofilm and c-di-GMP production, there was no significant change between ΔopgGH-ΔrcsF/ΔopgGH-ΔrcsC/ΔopgGH-ΔrcsB double mutant and ΔrcsF/ΔrcsC/ΔrcsB single mutant in terms of biofilm formation and intracellular c-di-GMP concentration (P > 0.05) (Figures 4, 5), indicating that loss of OPGs will not affect the biofilm and c-di-GMP production in the absence of Rcs phosphorelay in Y. enterocolitica.
However, disruption of the Rcs phosphorelay led to the smaller cell size of Y. enterocolitica. The ΔrcsF, ΔrcsC, and ΔrcsB single mutants were 24, 17, and 27% smaller, respectively, compared to the wild-type strain (Figure 6 and Supplementary Figure 4). Thus the cell size of the double mutant strains was further reduced compared with the ΔopgGH single mutant. The cell size of the ΔopgGH-ΔrcsF, ΔopgGH-ΔrcsC, and ΔopgGH-ΔrcsB double mutants were 17, 12, and 23%, respectively, smaller than those of the ΔopgGH single mutant (Figure 6 and Supplementary Figure 4).
Relationship Between OPGs and the Rcs Phosphorelay According to the Expression of RcsB-Regulated Genes in Y. enterocolitica
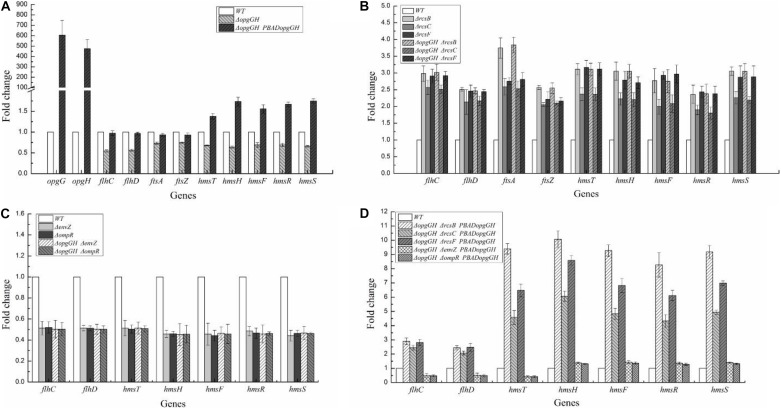
The above data indicate that the reduced motility, biofilm formation and c-di-GMP production caused by OPGs deficiency could be restored by disruption of the Rcs phosphorelay. Furthermore, OPGs deficiency could not affect these phenotypes in the absence of Rcs phosphorelay in Y. enterocolitica. These results provide evidence that loss of OPGs synthesis decreased Y. enterocolitica motility, biofilm formation and c-di-GMP production through acting on the Rcs phosphorelay. In D. dadantii, the Rcs phosphorelay regulates the flhDC and ftsAZ operons required for the expression of flagellar apparatus genes and cell division, respectively (Bontemps-Gallo et al., 2013). Furthermore, Rcs signaling controls the transcription of genes involved in biofilm formation: hmsT encoding diguanylate cyclase essential for c-di-GMP biosynthesis and hmsHFRS required for biosynthesis of poly-β-1,6-N-acetylglucosamine exopolysaccharide (EPS) (Fang et al., 2015). In this study, we analyzed transcription levels of these genes to determine whether OPGs deficiency activated the Rcs phosphorelay in Y. enterocolitica. The results revealed that all these genes were repressed in the absence of OPGs biosynthesis. Thus, the expression of flhDC, hmsT, hmsHFRS, and ftsAZ in the ΔopgGH strain was downregulated by an average of 45, 33, 34, and 27%, respectively, but the expression of all these genes in the ΔopgGH-PBADopgGH strain was restored when compared to the wild-type strain; furthermore, the levels of hmsT and hmsHFRS exceeded those in the wild-type strain by an average of 1.38- and 1.68-fold (Figure 7A).
FIGURE 7.
Transcriptional changes in the mutant strains of Y. enterocolitica. Y. enterocolitica grown to the mid-log phase in LNNS medium and total RNA was extracted. Expression of opgGH, flhDC, ftsAZ, hmsHFRS, and hmsT were determined by RT-qPCR in vitro. The 16S rRNA gene was used as a normalization control. (A) Gene transcription in ΔopgGH and ΔopgGH-PBADopgGH strains. (B) Gene transcription in ΔrcsF, ΔrcsC, ΔrcsB, ΔopgGH-ΔrcsF, ΔopgGH-ΔrcsC, and ΔopgGH-ΔrcsB mutant strains. (C) Gene transcription in ΔenvZ, ΔompR, ΔopgGH-ΔenvZ, and ΔopgGH-ΔompR mutant strains. (D) Gene transcription in ΔopgGH-ΔrcsF-PBADopgGH, ΔopgGH-ΔrcsC-PBADopgGH, ΔopgGH-ΔrcsB-PBADopgGH, ΔopgGH-ΔenvZ-PBADopgGH, and ΔopgGH-ΔompR-PBADopgGH strains. The data are presented as the mean ± SD of at least three independent experiments.
However, expression levels of genes downregulated by OPGs deficiency were increased in all double-mutant strains. The transcription of flhDC, hmsT, hmsHFRS, and ftsAZ in ΔopgGH-ΔrcsF/ΔopgGH-ΔrcsC/ΔopgGH-ΔrcsB was upregulated by an average of 2.68-fold/2.34-fold/2.74-fold, 3.11-fold/2.36-fold/3.10-fold, 2.74-fold/2.07-fold/2.81-fold, and 2.48-fold/2.32-fold/3.19-fold, respectively, when compared to the wild-type strain (Figure 7B). The deletion of rcsF/rcsC/rcsB also resulted in increase in these genes expression, however, there was no significant change between ΔopgGH-ΔrcsF/ΔopgGH-ΔrcsC/ΔopgGH-ΔrcsB double mutant and ΔrcsF/ΔrcsC/ΔrcsB single mutant in the transcription of flhDC, hmsT, hmsHFRS, and ftsAZ (P > 0.05) (Figure 7B). All these results indicated that that deletion of any Rcs phosphorelay gene reversed transcriptional changes caused by the lack of OPGs.
Disruption of the EnvZ/OmpR Phosphorelay Further Reduced Motility, c-di-GMP Production, and Biofilm Formation in the Y. enterocolitica ΔopgGH Mutant
It was shown that defects in motility caused by OPGs deficiency could be restored by disrupting the envZ-ompR operon in E. coli (Fiedler and Rotering, 1988). In this study, we constructed ΔopgGH-ΔenvZ and ΔopgGH-ΔompR double mutants to test the effect of EnvZ/OmpR phosphorelay on motility and flhDC expression in the Y. enterocolitica ΔopgGH mutant. However, as described previously, OmpR directly, positively regulates the expression of flhDC in Y. enterocolitica, as well as in Y. pseudotuberculosis (Hu et al., 2009; Raczkowska et al., 2011; Meng et al., 2019). The swim diameter and flhDC expression decreased by 47 and 49%, respectively, due to the inactivation of envZ or ompR (Figures 3, 7C). Thus both the ΔopgGH-ΔenvZ and ΔopgGH-ΔompR strains reduced swim motility and flhDC expression by 46 and 50%, respectively, when compared to the wild-type strain (Figures 3, 7C). In contrast to E. coli, disruption of the EnvZ/OmpR phosphorelay in Y. enterocolitica did not recover motility decreased by OPGs deficiency.
In addition, the results revealed that inactivation of envZ or ompR markedly decreased both biofilm formation and c-di-GMP production in the ΔopgGH strain. After 72 h of incubation, both ΔopgGH-ΔenvZ and ΔopgGH-ΔompR double mutants were decreased by 21% in biofilm formation when compared to the ΔopgGH single mutant (Figure 4). The intracellular c-di-GMP production in ΔopgGH-ΔenvZ and ΔopgGH-ΔompR double mutants were 54 and 56% reduction, respectively, compared to the ΔopgGH single mutant (Figure 5). Correspondingly, the transcription of the hmsT and hmsHFRS were downregulated by an average of 50 and 54% when compared to the wild-type strain (Figure 7C).
Rcs and EnvZ/OmpR Phosphorelays Showed Opposite Effects on OPGs-Induced Biofilm and c-di-GMP Production in Y. enterocolitica
In this study, it was shown that Y. enterocolitica was capable of forming biofilms in LNNS medium and the increase in OPGs greatly induced biofilm formation and c-di-GMP production (Figures 4, 5 and Supplementary Figure 3). Since the inactivation of Rcs phosphorelay upregulated the flhDC, hmsT, and hmsHFRS expression which is responsible for biofilm and c-di-GMP production (Figure 7B), we further investigated the effect of Rcs phosphorelay on OPGs-induced biofilm formation and c-di-GMP synthesis by transfecting double mutants with pBAD24-opgGH to create ΔopgGH-ΔrcsF-PBADopgGH, ΔopgGH-ΔrcsC-PBADopgGH and ΔopgGH-ΔrcsB-PBADopgGH strains. As a result, the ΔopgGH-ΔrcsF-PBADopgGH, ΔopgGH-ΔrcsC-PBADopgGH, and ΔopgGH-ΔrcsB-PBADopgGH strains had increased swim diameter by 31, 12, and 35%, respectively, compared to the ΔopgGH-PBADopgGH strain (Figure 3). The biofilm formation in ΔopgGH-ΔrcsF-PBADopgGH, ΔopgGH-ΔrcsC-PBADopgGH, and ΔopgGH-ΔrcsB-PBADopgGH strains were 86, 68, and 1.15-fold increase, respectively (Figure 4); correspondingly, in these strains, the c-di-GMP production was increased by 96, 48% and 1.27-fold when compared to the ΔopgGH-PBADopgGH strain (Figure 5). The expression of the flhDC, hmsT, and hmsHFRS genes in Δopg GH-ΔrcsF-PBADopgGH/ΔopgGH-ΔrcsC-PBADopgGH/ΔopgGH-ΔrcsB-PBADopgGH was upregulated by an average of 2.64-fold/2.26-fold/2.68-fold, 6.48-fold/4.58-fold/9.39-fold, and 7.13-fold/5.04-fold/9.20-fold, respectively, when compared to the wild-type strain (Figure 7D). These results indicated that disruption of the Rcs phosphorelay increased motility and further promoted induction of biofilm and c-di-GMP production by OPGs in ΔopgGH-PBADopgGH strain.
We also investigated the relationship between OPGs overproduction and the EnvZ/OmpR phosphorelay using the same approach, i.e., transforming ΔopgGH-ΔenvZ and ΔopgGH-ΔompR double mutants with pBAD24-opgGH. Although OPGs overproduction upregulated c-di-GMP and biofilm production in ΔopgGH-ΔenvZ-PBADopgGH and ΔopgGH-ΔompR-PBADopgGH strains compared to the double mutants, these effects were significantly weaker than those in the ΔopgGH-PBADopgGH strain (Figures 4, 5 and Supplementary Figure 3). Both the ΔopgGH-ΔenvZ-PBADopgGH and ΔopgGH-ΔompR-PBADopgGH strains had decreased swim diameter by 52% compared to the ΔopgGH-PBADopgGH strain (Figure 3). The biofilm formation was also decreased by 39 and 38%, respectively (Figure 4); accordingly, the c-di-GMP production was decreased by 58 and 57% when compared to the ΔopgGH-PBADopgGH strain (Figure 5). The expression of the flhDC and hmsT genes in ΔopgGH-ΔenvZ-PBADopgGH/ΔopgGH-ΔompR-PBADopgGH strains was downregulated by an average of 50%/51% and 58%/59%, respectively, when compared to the wild-type strain (Figure 7D). Though OPGs overproduction upregulated the hmsHFRS expression in ΔopgGH-ΔenvZ-PBADopgGH and ΔopgGH-ΔompR-PBADopgGH strains compared to the wild-type strain, the expression was lower than those in the ΔopgGH-PBADopgGH strain (Figures 7A,D). All these results indicated that the disruption of the EnvZ/OmpR phosphorelay decreased motility and prevented the induction of biofilm and c-di-GMP production by OPGs in ΔopgGH-PBADopgGH strain.
Discussion
Osmoregulated periplasmic glucans are important signaling molecules existing in the periplasmic space of many Gram-negative bacteria, where they participate in environmental perception and regulation of genes involved in virulence of Enterobacteriaceae pathogens (Bontemps-Gallo and Lacroix, 2015; Bontemps-Gallo et al., 2017). However, the role of OPGs in Y. enterocolitica was previously unknown. Results in this study indicate that deletion of opgGH operon in the chromosome of Y. enterocolitica decreased motility, c-di-GMP production, biofilm formation and cell size of the bacteria, indicating that OPGs regulate several functions and confer growth advantage to Y. enterocolitica. In addition to the presence in the periplasmic space, OPGs are also secreted into the extracellular space, where they can directly interact with antibiotics, thus preventing their cell-damaging effects (Bontemps-Gallo and Lacroix, 2015). Previous findings suggest that OPGs are critical for the tolerance of P. aeruginosa to aminoglycoside antibiotics by sequestering antibiotic molecules away from their cellular targets only when the bacteria were in a biofilm (Mah et al., 2003). In this study, the effect of OPGs on antibiotic sensitivity was performed in planktonic-grown Y. enterocolitica, and it was found that the OPGs of wild-type strain did not confer resistance to antibiotics, which was similar to that of in P. aeruginosa. However, the overexpression of opgGH resulted in more than twofold increase in OPGs production compared to the wild-type strain (Figure 2B), which enhanced Y. enterocolitica tolerance to chloramphenicol and polymyxin B with different mechanisms of action. Considering that OPGs can be secreted extracellularly, it can be easily assumed that the ΔopgGH-PBADopgGH strain produced so much OPGs, leading to a leakage of OPGs into the external environment. Thus, the phenotype observed is the consequence of OPGs overproduction but does not indicate that OPGs are involved in antibiotic resistance in Y. enterocolitica.
Recent study reported that Y. pestis lost the opgGH operon during its emergence from the enteropathogen Y. pseudotuberculosis. Y. pseudotuberculosis did not produce OPGs even if the opgGH operon was expressed. Inactivation of the opgGH operon showed normal motility, biofilm formation, resistance to polymyxin B and virulence (Quintard et al., 2015). Furthermore, Y. pestis re-introducing opgGH grew normally and was able to complete its infectious cycle (flea-host) (Quintard et al., 2015). Data in this study showed that OPGs can be detected in Y. enterocolitica under low-nutrient low-salt conditions and that deletion of the opgGH operon generated a pleiotropic phenotype and OPGs overproduction conferred Y. enterocolitica higher resistance to polymyxin B. NCBI homologous alignment revealed that opgG and opgH nucleotide between the two species shows 81.75 and 80.26% identities over the whole sequence respectively, indicating the difference in nucleotide sequences of opgGH between the two species, which help to explain why opgGH in Y. enterocolitica encodes functional OPGs, but opgGH in Y. pseudotuberculosis had no effect of this. Or as explained by Quintard et al. (2015) that Y. pseudotuberculosis might produce OPGs under particular conditions or might lack an essential factor for OPGs biosynthesis, since the opgGH operon from Y. pseudotuberculosis could encode functional OPGs in E. coli and D. dadantii (Quintard et al., 2015). However, inactivation of opgGH reduced the cell size in both Y. enterocolitica and Y. pseudotuberculosis.
The Rcs phosphorelay is an atypical TCS conserved in Enterobacteriaceae that allows bacteria to perceive external environment stimuli and modify gene expression (Clarke, 2010; Guo and Sun, 2017); however, its functional activity in Y. enterocolitica has not been reported. In the present work, we found that the flhDC, hmsT, hmsHFRS, and ftsAZ genes expression regulated by Rcs phosphorelay were repressed by OPGs deficiency, but deletion of rcsF, rcsC, and rcsB encoding Rcs phosphorelay components in ΔopgGH mutant restored expression levels of flhDC, hmsT, hmsHFRS and ftsAZ, as well as cell motility, c-di-GMP production and biofilm formation in Y. enterocolitica. In fact, the increased flhDC, hmsT, hmsHFRS, and ftsAZ were also observed in ΔrcsF, ΔrcsC, and ΔrcsB single mutants cultured in LNNS medium, suggesting that in Y. enterocolitica, the Rcs system is activated under low-nutrient low-salt conditions, which may explain why deletion of rcsF/rcsC/rcsB not only rescued but further promoted motility, c-di-GMP production and biofilm formation in the ΔopgGH mutant. However, there was no significant difference between ΔopgGH-ΔrcsF/ΔopgGH-ΔrcsC/ΔopgGH-ΔrcsB double mutant and ΔrcsF/ΔrcsC/ΔrcsB single mutant in terms of the expression level of all these genes, suggesting that OPGs deficiency could not affect these genes’ expression in the inactivation of Rcs phosphorelay in Y. enterocolitica. All these results provide evidence that downregulation of OPGs synthesis decreases Y. enterocolitica motility, c-di-GMP production and biofilm formation through activation of the Rcs phosphorelay. In addition, it has been reported that the ΔopgGH mutant decreased swim diameter by 70% and lost the capacity to form a biofilm in D. dadantii (Bouchart, 2006; Bontemps-Gallo et al., 2013), while in this study the swim diameter and biofilm formation only decreased by 21 and 24%, respectively, in LNNS medium but the results can be reproduced. It was suspected that this small change may be related to the over-activation of the Rcs system in the wild-type of Y. enterocolitica under low-nutrient low-salt conditions, thus even if the OPGs deficiency leads to activation of the Rcs system, the phenotypic changes in the response were not so obvious.
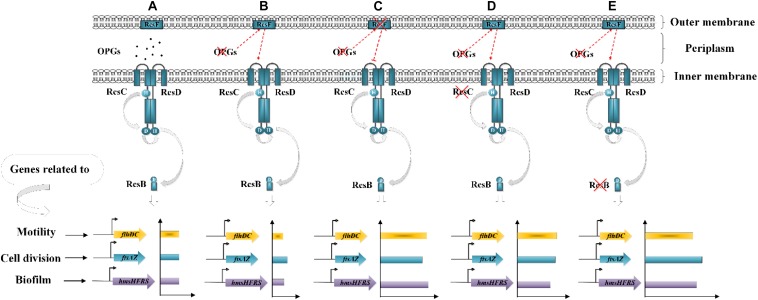
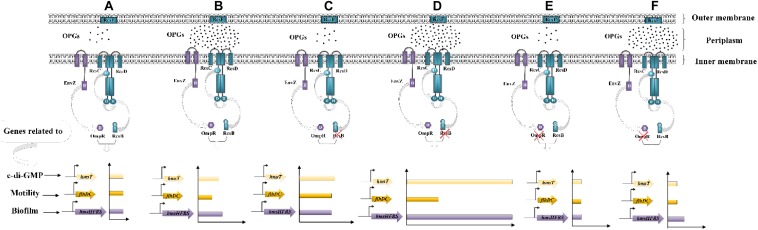
Based on the expression of RcsB-regulated operons, we propose a model linking OPGs to the Rcs phosphorelay in Y. enterocolitica (Figure 8). Combined the previously reported state of phosphorylation of RcsB in the wild-type of D. dadantii (Madec et al., 2014), we can propose that: in the wild-type Y. enterocolitica (Figure 8A), OPGs are synthesized and RcsB is phosphorylated at a certain level providing certain repression of flhDC, hmsHFRS and ftsAZ, whereas in the absence of OPGs synthesis (Figure 8B), the activation of the Rcs phosphorelay is induced, leading to decrease in motility, biofilm formation and cell division through downregulation of flhDC, hmsHFRS, and ftsAZ. In the absence of both OPGs and Rcs signaling (Figures 8C–E), when RcsB is not phosphorylated, motility, biofilm formation and cell division are restored through upregulation of flhDC, hmsHFRS, and ftsAZ. This model characterizes the relationship between OPGs and the Rcs phosphorelay illustrating an important role of OPGs in Y. enterocolitica, which regulate the Rcs signal transduction pathway, preventing its overactivation, thus promoting cell motility, division, and biofilm formation. In this study, OPGs deficiency could not affect the expression of these genes in the absence of RcsF, indicating that the RcsF protein is required for the perception of OPGs defect in Y. enterocolitica. It should be noted that RcsF-dependent activation of the Rcs phosphorelay is a consequence of BamA (the core protein for β-barrel assembly) failing to bind RcsF and funnel it to OmpA (the β-barrel) (Cho et al., 2014). However, it still remains to be elucidated whether this mechanism works for Y. enterocolitica when Rcs system is activated by loss of OPGs.
FIGURE 8.
Working model of the relationship between OPGs and the Rcs phosphorelay in Y. enterocolitica. (A) In the wild-type strain, the Rcs system represses cell motility, division and biofilm formation by inhibiting flhDC, ftsAZ, and hmsHFRS expression. (B) OPGs deficiency activates the Rcs phosphorelay and inhibits flhDC, ftsAZ, and hmsHFRS expression, resulting in further repression of cell motility, division, and biofilm formation. (C–E) Disruption of the Rcs phosphorelay pathway downregulates RcsB phosphorylation, restoring cell motility, division, and biofilm formation by enhancing the expression of flhDC, ftsAZ, and hmsHFRS.
In Y. enterocolitica, ΔrcsB single mutant upregulated the ftsAZ operon, indicating that the ftsAZ genes are repressed by RcsB, which is in contrast with previous findings that the ftsAZ operon was activated by RcsB in E. coli and Proteus mirabilis (Howery et al., 2016). Our finding that activation of the Rcs system decreased ftsAZ expression is different from earlier reports showing that Rcs activation enhanced ftsAZ expression in D. dadantii (Bontemps-Gallo et al., 2013) and E. coli (Hill et al., 2013). These data suggest the existence of species-specific differences in the regulation patterns of the Rcs system. Previous studies have reported that OpgH regulates bacterial cell architecture by sequestering FtsZ and, thus, affecting cell length independently from OPGs synthesis (Hill et al., 2013; Quintard et al., 2015). Here, we observed reduction of ftsAZ expression and decrease in the cell size of the ΔopgGH mutant, which may be due not to the lack of OPGs or activation of the Rcs system but rather to the absence of other OpgH activities unrelated to OPGs biosynthesis. However, as FtsZ levels should be increased by about 50% to cause cell size reduction of 15–20% (Hill et al., 2013), it may be speculated that a possible reason for cell size decrease in ΔopgGH-ΔrcsF, ΔopgGH-ΔrcsC, and ΔopgGH-ΔrcsB strains could be the overexpression of ftsAZ (Figure 7B).
It was reported that OmpR negatively regulates the expression of flhDC in E. coli, thus defects in motility caused by OPGs deficiency could be restored by disrupting the EnvZ/OmpR system (Fiedler and Rotering, 1988). While in Y. enterocolitica, OmpR was shown to positively regulate the expression of flhDC and motility (Raczkowska et al., 2011; Meng et al., 2019). Thus, inactivation of envZ or ompR in Y. enterocolitica ΔopgGH mutant led to 46% reduction in swim motility compared to the wild-type strain, which is in agreement with a recent report in D. dadantii that inactivation of envZ or ompR in the opgG background resulted in a 40% reduction in motility relative to the wild-type strain (Caby et al., 2018). Unlike E. coli, inactivation of EnvZ/OmpR phosphorelay system cannot restore flhDC expression and motility in the OPGs-deficient strain both in Y. enterocolitica and D. dadantii, Rcs and EnvZ/OmpR phosphorelays showed the opposite effects on the regulation of flhDC expression and motility in Y. enterocolitica and D. dadantii. In addition, it should be noted that Rcs and EnvZ/OmpR systems control the mechanism of OPGs succinylation in D. dadantii, but, unlike for Rcs system, EnvZ/OmpR system is not controlled by OPGs concentration but requires OPGs for proper activation (Bontemps-Gallo et al., 2016; Caby et al., 2018).
The ability to form biofilms is important for protection of bacteria from various stresses and critically depends on their motility (Brzostek et al., 2012; Pruss, 2017). In this work, we found that Y. enterocolitica was capable of forming biofilms under low-nutrient low-salt conditions and that OPGs could play an important role in promoting the process (Figure 4 and Supplementary Figure 3). The flhDC operon encodes the master regulator of flagellum biosynthesis and is the key factor in the initial attachment of biofilms (Raczkowska et al., 2011). The hmsHFRS operon is responsible for the synthesis of poly-β-1,6-N-acetylglucosamine EPS constituting the extracellular polymeric substance, which is a key factor in promoting biofilm maturation (Bomchil et al., 2003; Fang et al., 2015). The hmsT encodes diguanylate cyclase required for biosynthesis of c-di-GMP, a second messenger involved in the regulation of a variety of bacterial behaviors, including biofilm formation (Fang et al., 2015). In this study, analysis of flhDC, hmsHFRS, and hmsT expression revealed a mechanism underlying the effect of Rcs and EnvZ/OmpR phosphorelays on OPGs-induced biofilm formation (Figure 9). OPGs overproduction in the periplasm promoted biofilm and c-di-GMP production in the presence of both phosphorelays by inducing the expression of hmsHFRS and hmsT genes (Figure 9B). Inactivation of the Rcs phosphorelay upregulated the flhDC, hmsHFRS, and hmsT expression, leading to the increased motility and further promoted the induction of biofilm and c-di-GMP production regulated by OPGs (Figure 9D); in contrast, inactivation of the EnvZ/OmpR phosphorelay downregulated the flhDC, hmsHFRS and hmsT expression, resulting in decreased motility and prevented the induction of biofilm and c-di-GMP production regulated by OPGs (Figure 9F). Thus, Rcs and EnvZ/OmpR phosphorelays exert the opposite effects on the regulation of OPGs-induced c-di-GMP and biofilm production in Y. enterocolitica. A recent study showed that Pantoea alhagi senses environmental osmolarity changes through the EnvZ/OmpR system and LrhA to regulate synthesis of OPGs, EPS production and flagella-dependent motility. OPGs control the Rcs activation in a concentration-dependent manner, and a high concentration of OPGs induced by increased medium osmolarity is maintained to achieve the high level of activation of the Rcs phosphorelay which results in enhanced EPS synthesis and decreased motility in P. alhagi (Li et al., 2019). As motility and biofilm formation are critical for bacterial colonization of the host, these findings indicate that OPGs associated with Rcs and EnvZ/OmpR systems play a key role in shaping the pathogenic phenotype of bacteria. Further studies are required to fully understand the mechanism underlying OPGs-mediated biofilm formation in Y. enterocolitica, which is likely multi-factorial.
FIGURE 9.
Effects of Rcs and EnvZ/OmpR phosphorelays on OPGs-induced biofilm and c-di-GMP production in Y. enterocolitica. (A,B) OPGs overproduction induces the expression of hmsHFRS and hmsT, thus promoting biofilm and c-di-GMP production in the presence of both Rcs and EnvZ/OmpR phosphorelays. (C,D) Disruption of the Rcs phosphorelay upregulates the expression of flhDC, hmsHFRS and hmsT, resulting in increased motility and further promoted the induction of biofilm and c-di-GMP production regulated by OPGs. (E,F) Disruption of the EnvZ/OmpR phosphorelay decreases flhDC, hmsHFRS, and hmsT expression, inhibiting motility and preventing the induction of biofilm and c-di-GMP production regulated by OPGs.
Conclusion
This study shows that OPGs are a part of the Rcs signal transduction pathway in Y. enterocolitica, regulating gene expression and producing a pleiotropic phenotype by disturbing the Rcs system. OPGs enhance antibiotic resistance of Y. enterocolitica; furthermore, they promote cell motility, c-di-GMP biosynthesis and biofilm production in low-nutrient low-salt conditions. It was also found that Rcs and EnvZ/OmpR phosphorelays exert the opposite effects on the regulation of OPGs-induced c-di-GMP and biofilm production in Y. enterocolitica. These findings reveal part of the biological function of OPGs in Y. enterocolitica and its relationship with Rcs and EnvZ/OmpR systems, which is important for understanding complex mechanisms underlying Y. enterocolitica pathogenicity.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
JM performed the experiments under the guidance of JC. JC and JM developed the idea for the study and designed the research. JM, CH, and XH analyzed the experimental data and drafted the manuscript. DL and BH made substantial contributions to conception, interpretation of data, and revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the members of the JC laboratory for helpful discussions. We are grateful to Jiaqi Bai and Junhong Xu for critically reading the manuscript and providing helpful comments.
Footnotes
Funding. This work was supported by National Natural Science Foundation of China (31671830) and National Key R&D Program of China (2017YFC1600806).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00122/full#supplementary-material
References
- Bang I. S., Kim B. H., Foster J. W., Park Y. K. (2000). OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar typhimurium. J. Bacteriol. 182 2245–2252. 10.1128/JB.182.8.2245-2252.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari M. L., Hossain M. A., Isshiki K., Ukuku D. (2011). Behavior of Yersinia enterocolitica in foods. J. Pathog. 2011:420732. 10.4061/2011/420732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat A. A., Jun W., Liu L., Kannan P., Dharne M., Pheh B., et al. (2009). Osmoregulated periplasmic glucans of Salmonella enterica serovar typhimurium are required for optimal virulence in mice. Microbiology 155 229–237. 10.1099/mic.0.023747-0 [DOI] [PubMed] [Google Scholar]
- Bomchil N., Watnick P., Kolter R. (2003). Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185 1384–1390. 10.1128/JB.185.4.1384-1390.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontemps-Gallo S., Bohin J. P., Lacroix J. M. (2017). Osmoregulated periplasmic glucans. EcoSal Plus 7 10.1128/ecosalplus.ESP-0001-2017 [DOI] [PubMed] [Google Scholar]
- Bontemps-Gallo S., Lacroix J. M. (2015). New insights into the biological role of the osmoregulated periplasmic glucans in pathogenic and symbiotic bacteria. Environ. Microbiol. Rep. 7 690–697. 10.1111/1758-2229.12325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontemps-Gallo S., Madec E., Dondeyne J., Delrue B., Robbe-Masselot C., Vidal O., et al. (2013). Concentration of osmoregulated periplasmic glucans (OPGs) modulates the activation level of the RcsCD RcsB phosphorelay in the phytopathogen bacteria Dickeya dadantii. Environ. Microbiol. 15 881–894. 10.1111/1462-2920.12054 [DOI] [PubMed] [Google Scholar]
- Bontemps-Gallo S., Madec E., Robbe-Masselot C., Souche E., Dondeyne J., Lacroix J. M. (2016). The opgC gene is required for OPGs succinylation and is osmoregulated through RcsCDB and EnvZ/OmpR in the phytopathogen Dickeya dadantii. Sci. Rep. 6:19619. 10.1038/srep19619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone E. J. (1997). Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10 257–276. 10.1128/CMR.10.2.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchart F. (2006). Impact des Mutations OPG chez Erwinia Chrysanthemi: Recherche de Suppresseurs et Analyse Protéomique des Mutants, Impact of OPG Mutations of Erwinia Chrysanthemi. Ph.D. thesis, University of Lille, Villeneuve d’Ascq. [Google Scholar]
- Bouchart F., Boussemart G., Prouvost A. F., Cogez V., Madec E., Vidal O., et al. (2010). The virulence of a Dickeya dadantii 3937 mutant devoid of osmoregulated periplasmic glucans is restored by inactivation of the RcsCD-RcsB phosphorelay. J. Bacteriol. 192 3484–3490. 10.1128/JB.00143-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briones G., De Iannino N. I., Roset M., Vigliocco A., Paulo P. S., Ugalde R. A. (2001). Brucella abortus cyclic beta-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect. Immun. 69 4528–4535. 10.1128/IAI.69.7.4528-4535.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzostek K., Brzostkowska M., Bukowska I., Karwicka E., Raczkowska A. (2007). OmpR negatively regulates expression of invasin in Yersinia enterocolitica. Microbiology 153 2416–2425. 10.1099/mic.0.2006/003202-0 [DOI] [PubMed] [Google Scholar]
- Brzostek K., Raczkowska A., Zasada A. (2003). The osmotic regulator OmpR is involved in the response of Yersinia enterocolitica O: 9 to environmental stresses and survival within macrophages. FEMS Microbiol. Lett. 228 265–271. 10.1016/s0378-1097(03)00779-1 [DOI] [PubMed] [Google Scholar]
- Brzostek K., Skorek K., Raczkowska A. (2012). OmpR, a central integrator of several cellular responses in Yersinia enterocolitica. Adv. Exp. Med. Biol. 954 325–334. 10.1007/978-1-4614-3561-7_40 [DOI] [PubMed] [Google Scholar]
- Caby M., Bontemps-Gallo S., Gruau P., Defrue B., Madecl E., Lacroix J. M. (2018). The EnvZ-OmpR two-component signaling system is inactivated in a mutant devoid of osmoregulated periplasmic glucans in Dickeya dadantii. Front. Microbiol. 9:2459. 10.3389/fmicb.2018.02459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanie-Cornet M. P., Cam K., Jacq A. (2006). RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J. Bacteriol. 188 4264–4270. 10.1128/JB.00004-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. H., Szewczyk J., Pesavento C., Zietek M., Banzhaf M., Roszczenko P., et al. (2014). Detecting envelope stress by monitoring β-barrel assembly. Cell 159 1652–1664. 10.1016/j.cell.2014.11.045 [DOI] [PubMed] [Google Scholar]
- Clarke D. J. (2010). The Rcs phosphorelay more than just a two-component pathway. Future Microbiol. 5 1173–1184. 10.2217/FMB.10.83 [DOI] [PubMed] [Google Scholar]
- Clarke E. J., Voigt C. A. (2011). Characterization of combinatorial patterns generated by multiple two-component sensors in E. coli that respond to many stimuli. Biotechnol. Bioeng. 108 666–675. 10.1002/bit.22966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2012). Performance Standards for Antimicrobial Disk Susceptibility Tests, Approved Standard- Eleventh Edition (Wayne, PA: Clinical and Laboratory Standards Institute; ), M02–M11. [Google Scholar]
- Dorrell N., Li S. R., Everest P. H., Dougan G., Wren B. W. (1998). Construction and characterisation of a Yersinia enterocolitica O:8 ompR mutant. FEMS Microbiol. Lett. 165 145–151. 10.1111/j.1574-6968.1998.tb13139.x [DOI] [PubMed] [Google Scholar]
- Fabrega A., Vila J. (2012). Yersinia enterocolitica: pathogenesis, virulence and antimicrobial resistance. Enferm. Infecc. Microbiol. Clin. 30 24–32. 10.1016/j.eimc.2011.07.017 [DOI] [PubMed] [Google Scholar]
- Fang N., Yang H. Y., Fang H. H., Liu L., Zhang Y. Q., Wang L., et al. (2015). RcsAB is a major repressor of Yersinia biofilm development through directly acting on hmsCDE, hmsT, and hmsHFRS. Sci. Rep. 5:9566. 10.1038/srep09566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler W., Rotering H. (1988). Properties of Escherichia coli mutants lacking membrane-derived oligosaccharides. J. Biol. Chem. 263 14684–14689. [PubMed] [Google Scholar]
- Girgis H. S., Liu Y. C., Ryu W. S., Tavazoie S. (2007). A comprehensive genetic characterization of bacterial motility. PLoS Genet. 3:e0030154. 10.1371/journal.pgen.0030154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. P., Sun Y. C. (2017). New insights into the non-orthodox two component rcs phosphorelay system. Front. Microbiol. 8:2014. 10.3389/fmicb.2017.02014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill N. S., Buske P. J., Shi Y., Levin P. A. (2013). A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet. 9:e1003663. 10.1371/journal.pgen.1003663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howery K. E., Clemmer K. M., Rather P. N. (2016). The Rcs regulon in Proteus mirabilis: implications for motility, biofilm formation, and virulence. Curr. Genet. 62 775–789. 10.1007/s00294-016-0579-1 [DOI] [PubMed] [Google Scholar]
- Hu Y., Wang Y., Ding L., Lu P., Atkinson S., Chen S. (2009). Positive regulation of flhDC expression by OmpR in Yersinia pseudotuberculosis. Microbiology 155 3622–3631. 10.1099/mic.0.030908-0 [DOI] [PubMed] [Google Scholar]
- Huovinen E., Sihvonen L. M., Virtanen M. J., Haukka K., Siitonen A., Kuusi M. (2010). Symptoms and sources of Yersinia enterocolitica -infection: a case-control study. BMC Infect. Dis. 10:122. 10.1186/1471-2334-10-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. J., Young B. M., Young G. M. (2008). Effect of flagellar mutations on Yersinia enterocolitica biofilm formation. Appl. Environ. Microbiol. 74 5466–5474. 10.1128/AEM.00222-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liang H., Wei Z., Bai H., Li M., Li Q., et al. (2019). An osmoregulatory mechanism operating through OmpR and LrhA controls the motile-sessile switch in the plant growth-promoting bacterium Pantoea alhagi. Appl. Environ. Microbiol. 85:e00077-19. 10.1128/AEM.00077-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. X., Rose J., Huang S. J., Hu Y. B., Wu Q., Wang D., et al. (2017). A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases. Nat. Commun. 8:2104. 10.1038/s41467-017-02310-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madec E., Bontemps-Gallo S., Lacroix J. M. (2014). Increased phosphorylation of the RcsB regulator of the RcsCDB phosphorelay in strains of Dickeya dadantii devoid of osmoregulated periplasmic glucans revealed by Phos-tag gel analysis. Microbiology 160 2763–2770. 10.1099/mic.0.081273-0 [DOI] [PubMed] [Google Scholar]
- Mah T. F., Pitts B., Pellock B., Walker G. C., Stewart P. S., O’Toole G. A. (2003). A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426 306–310. 10.1038/nature02122 [DOI] [PubMed] [Google Scholar]
- Meng J., Bai J. Q., Xu J. H., Huang C., Chen J. Y. (2019). Differential regulation of physiological activities by RcsB and OmpR in Yersinia enterocolitica. FEMS Microbiol. Lett. 336:fnz210. 10.1093/femsle/fnz210 [DOI] [PubMed] [Google Scholar]
- Meng J., Wang B. Y., Liu D. Y., Tao C., Wang Z. W., Zhao X. M. (2016). High-yield anaerobic succinate production by strategically regulating multiple metabolic pathways based on stoichiometric maximum in Escherichia coli. Microb. Cell Fact. 15:141. 10.1186/s12934-016-0536-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page F., Altabe S., Hugouvieux-Cotte-Pattat N., Lacroix J. M., Robert-Baudouy J., Bohin J. P. (2001). Osmoregulated periplasmic glucan synthesis is required for Erwinia chrysanthemi pathogenicity. J. Bacteriol. 183 3134–3141. 10.1128/JB.183.10.3134-3141.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss B. M. (2017). Involvement of two-component signaling on bacterial motility and biofilm development. J. Bacteriol. 199:e00259-17. 10.1128/JB.00259-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintard K., Dewitte A., Reboul A., Madec E., Bontempsgallo S., Dondeyne J., et al. (2015). Evaluation of the role of the opgGH operon in Yersinia pseudotuberculosis and its deletion during the emergence of Yersinia pestis. Infect. Immun. 83 3638–3647. 10.1128/IAI.00482-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczkowska A., Skorek K., Bielecki J., Brzostek K. (2011). OmpR controls Yersinia enterocolitica motility by positive regulation of flhDC expression. Antonie Van Leeuwenhoek 99 381–394. 10.1007/s10482-010-9503-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Bonny T. S., Stonsaovapak S., Ananchaipattana C. (2011). Yersinia enterocolitica: epidemiological studies and outbreaks. J. Pathog. 2011 1–11. 10.4061/2011/239391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo F. D., Silhavy T. J. (1991). EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 222 567–580. 10.1016/0022-2836(91)90497-T [DOI] [PubMed] [Google Scholar]
- Schafer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145 69–73. 10.1016/0378-1119(94)90324-7 [DOI] [PubMed] [Google Scholar]
- Spiro R. G. (1966). Analysis of sugars found in glycoproteins. Methods Enzymol. 8 3–27. 10.1016/0076-6879(66)08005-4 [DOI] [Google Scholar]
- Thomson N. R., Howard S., Wren B. W., Holden M. T. G., Crossman L., Challis G. L., et al. (2006). The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e020206. 10.1371/journal.pgen.0020206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall E., Majdalani N., Gottesman S. (2018). The complex Rcs regulatory cascade. Annu. Rev. Microbiol. 72 111–139. 10.1146/annurev-micro-090817-062640 [DOI] [PubMed] [Google Scholar]
- Weissborn A. C., Rumley M. K., Kennedy E. P. (1992). Isolation and characterization of Escherichia coli mutants blocked in production of membrane-derived oligosaccharides. J. Bacteriol. 174 4856–4859. 10.1128/jb.174.14.4856-4859.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G. M., Miller V. L. (1997). Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol. Microbiol. 25 319–328. 10.1046/j.1365-2958.1997.4661829.x [DOI] [PubMed] [Google Scholar]
- Zadernowska A., Chajecka-Wierzchowska W., Laniewska-Trokenheim L. (2014). Yersinia enterocolitica: a dangerous, but often ignored, foodborne pathogen. Food Rev. Int. 30 53–70. 10.1080/87559129.2013.853775 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.