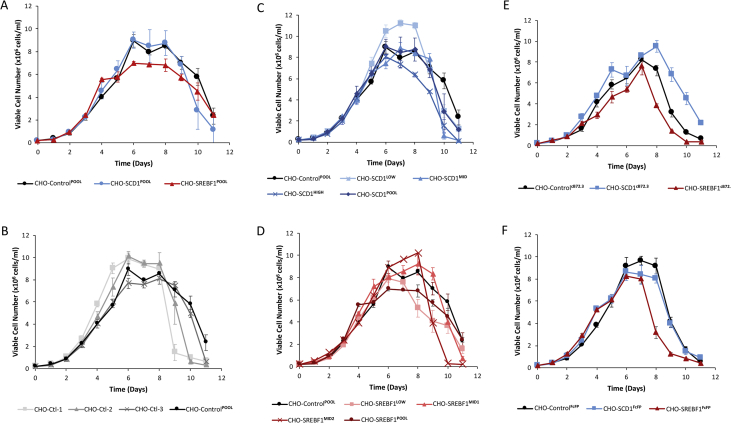
Fig. 2.
Growth profiles of CHOK1SV GS-KO™ cells engineered to overexpress lipid metabolism modifying genes SCD1 and SREBF1. Batch cultures were seeded at 0.2 × 106 viable cells/ml in a total culture volume of 20 ml and cell concentrations measured using a ViCell every 24 h. The CHO-ControlPOOL growth profile is shown in A-D for reference whilst lipid metabolism modified pool data are shown in (A), control monoclonal measurements are shown in (B), SCD1 monoclonal samples are shown in (C) and SREBF1 monoclonal measurements are shown in (D). Cell pools were constructed using lipid modified polyclonal cell pools as hosts such that they stably express either the cB72.3 monoclonal antibody or FcFP molecule. Growth profiles of these cells are shown in Figure (E) for cB72.3 expressing pools and Figure (F) for FcFP expressing pools. n = 3 for each data point and error bars show ± one standard deviation.