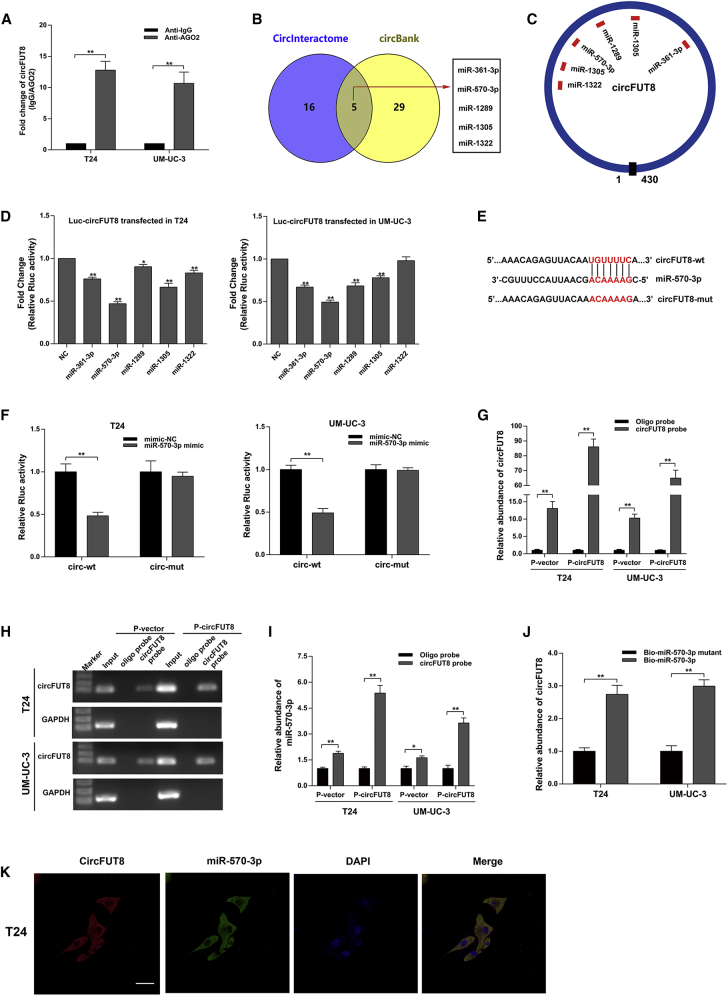
Figure 4.
circFUT8 Serves as a Sponge for miR-570-3p in BCa Cells
(A) The enrichment of circFUT8 was detected by quantitative real-time PCR after RIP assay for AGO2. (B) Potential target miRNAs of circFUT8 were predicted by CircInteractome and circBank. (C) Schematic diagram demonstrated the predicted binding sites of the five miRNAs in circFUT8. (D) Luciferase reporter assay: BCa cells transfected with psiCHECK-2-wild-type circFUT8 were co-transfected with the five miRNA mimics, respectively. (E and F) The characteristics of psiCHECK-2-wild-type circFUT8 (circ-wt) and psiCHECK-2 mutant-type circFUT8 (circ-mut) (E). Luciferase reporter assay indicated that miR-570-3p mimic significantly reduced the Rluc activity of circ-wt but not circ-mut (F). (G and H) circFUT8 was assessed by quantitative real-time PCR (G) after RNA pull-down assay, and the quantitative real-time PCR products from input and the RNA pull-down assay were used for gel electrophoresis (H). Oligo probe was used as negative control for RNA pull-down assay, and GAPDH was considered the internal control of gel electrophoresis. (I) miR-570-3p was analyzed by quantitative real-time PCR after RNA pull-down assay using circFUT8 probe. (J) circFUT8 captured by biotin-coupled miR-570-3p or miR-570-3p mutant mimics in T24 and UM-UC-3 cells was quantified by quantitative real-time PCR. (K) FISH assay demonstrates the co-localization of circFUT8 and miR-570-3p in the T24 cell line. Nuclei were stained with DAPI. The circFUT8 probe was labeled with Cy3, and the miR-570-3p probe was labeled with Cy5. Scale bar, 50 μm. Data are presented as the mean ± SEM of three experiments. *p < 0.05; **p < 0.01, Student’s t test.