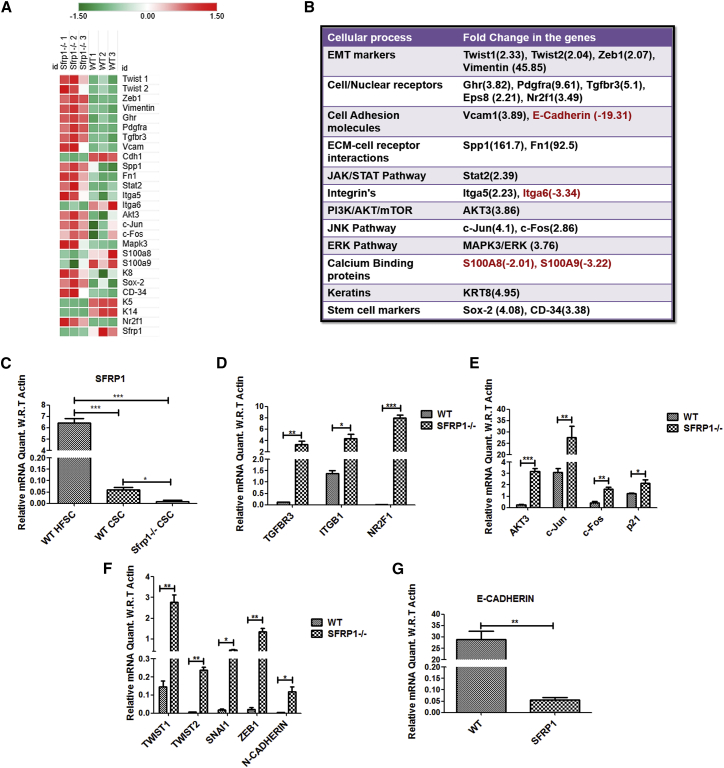
Figure 4.
Altered Signaling in Sfrp1−/− CSCs Compared with WT CSCs
(A) Heatmap of the significantly deregulated genes between WT CSCs and Sfrp1−/− CSCs (n = 3 biological replicates/genotype).
(B) Expression profile of various pathways in Sfrp1−/− CSCs compared with WT CSCs.
(C) Graph representing the expression levels of the Sfrp1 in WT HFSCs, WT CSCs, and Sfrp1−/− CSCs validated using quantitative real-time PCR (n = 3 biological replicates/genotype).
(D and E) Graphs representing the expression level changes in cell surface receptors and signaling molecules in WT CSCs and Sfrp1−/− CSCs validated using quantitative real-time PCR (n = 3 biological replicates/genotype).
(F and G) Graphs representing the expression level changes in EMT genes in WT CSCs and Sfrp1−/− CSCs validated using quantitative real-time PCR (n = 3 biological replicates/genotype).
FACS, fluorescence-activated cell sorting; HFSC, hair follicle stem cells; CSCs, cancer stem cells; EMT, epithelial to mesenchymal transition; WT, wild type; SFRP1−/−, homozygous knockout for Sfrp1. The mRNA expression levels were normalized to the expression of β-actin. Data were analyzed by Student's t test and presented as means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figures S3 and S4.