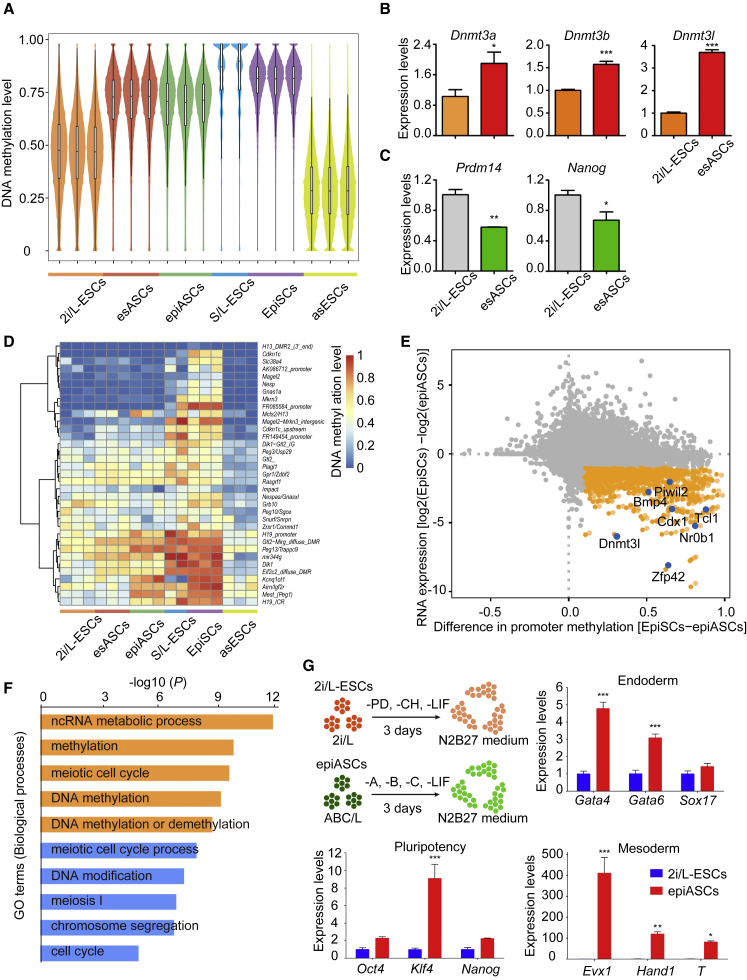
Figure 5.
Global Upregulation of DNA Methylation Level in esASCs and epiASCs
(A) Violin plot showing DNA methylation level of 2-kb genomic tiles. asESCs indicate that ASCs were cultured in 2i/L medium. Source data are provided in Table S1.
(B) Relative expression of DNA methyltransferase genes (Dnmt3a, Dnmt3b and Dnmt3l) measured by qPCR in esASCs. 2i/L-ESCs were used as control. Error bars indicate SEM (n = 3). Results were obtained from three independent experiments. p values were calculated by unpaired Student's t test, p < 0.05. (C) Relative expression of DNA methylation-associated genes (Prdm14 and Nanog) measured by qPCR in esASCs. 2i/L-ESCs were used as control. Error bars indicate SEM (n = 3). Results were obtained from three independent experiments. p values were calculated by unpaired Student's t test, p < 0.05.
(D) Heatmap showing DNA methylation level of ICRs in five different stem cells.
(E) Scatterplot of differential gene expression and difference in promoter DNA methylation level between EpiSCs and epiASCs. Genes upregulated in epiASCs with promoter demethylation are highlighted in orange. Source data are provided in Table S2.
(F) The top representative GO terms for Figure 5E upregulated and promoter demethylation genes.
(G) Experimental outline of ESCs differentiation. Mesoderm, endoderm, and pluripotency-associated genes were tested by qPCR, epiASCs were checked after 3 days induction, 2i/L-ESCs were used as control. Error bars indicate SEM (n = 3). Results were obtained from three independent experiments. p values were calculated by two-way ANOVA, p < 0.05.