Figure 1.
Reduction in Pol I Activity Extends Fruit Fly Lifespan
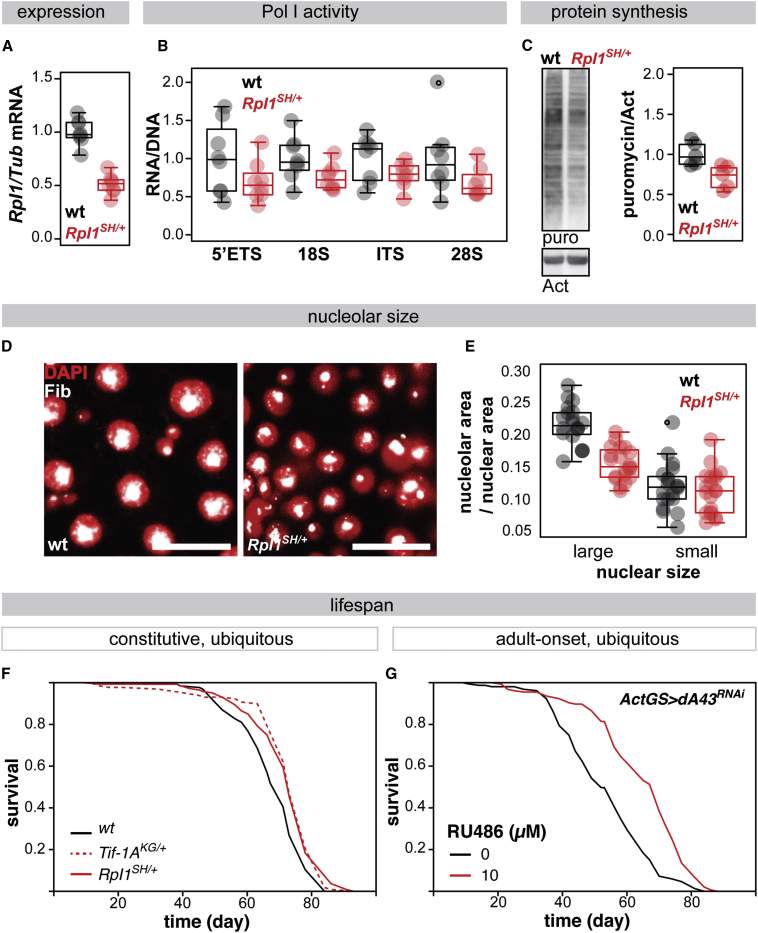
(A) Relative RpI1 transcript levels (n = 8 biologically independent samples, p < 10−4, t test).
(B) Ratio of RNA to DNA for the sequences present in pre-rRNA-rDNA (n = 8 biologically independent samples, effect of genotype p = 9 × 10−4, no significant effect of the target sequence or interaction, linear model [LM]).
(C) Relative levels of protein synthesis in whole flies determined by puromycin incorporation and western blotting, showing a representative blot (left) and quantification (right; n = 5 individual flies, p = 0.01, t test).
(D) Representative images of nucleoli (Fibrillarin staining) in posterior midguts (scale bar, 10 μm).
(E) Quantification of nucleolar size in large or small nuclei (area as proportion of nuclear area; n = 1–5 nucleoli per animal per nuclear size, 4–5 animals where values from the same animal are indicated as vertically aligned points in the boxplot, effect of genotype p = 0.01, nuclear size p < 1 × 10−4, interaction p = 2 × 10−4, mixed-effects LM with “animal” as random effect).
(F) Lifespans of RpI1SH/+ (n = 137 dead/15 censored flies), Tif-1AKG/+ (n = 118 dead/25 censored flies) and wild-type females (n = 118 dead/22 censored flies, p = 3 × 10−4 and 1 × 10−4, respectively, log-rank test).
(G) Lifespans of females after ubiquitous induction of dA43RNAi from day 2 of adulthood by RU486 feeding and controls (−RU486 n = 154 dead/4 censored flies, +RU486 n = 155 dead/1 censored flies, p = 4 × 10−12, log-rank test).
(A)–(E) were assessed in RpI1SH/+ and wild-type females. Boxplots show means and quintiles, with individual replicate points overlaid.
See also Figures S1 and S2.