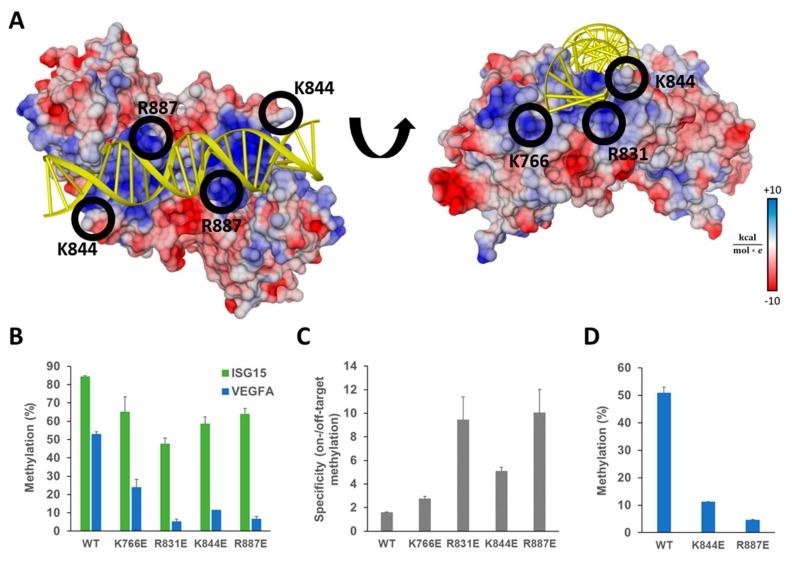
Figure 3.
Ab-3A3L containing DNMT3A mutants with reduced DNA binding shows lower off-target methylation but retains most of the on-target activity. (A) Crystal structure of the DNMT3A-DNA complex (pdb 5YX2, [36]) showing the DNA (yellow) bound to the central dimeric DNMT3A catalytic domain colored according to the surface charge (blue positive, red negative). Positively charged residues on the protein surface (blue patches) interact with the negatively charged DNA phosphodiester backbone. Amino acids K766, R831, K844, and R887 were selected for mutagenesis and replaced by glutamic acid to reduce the strength of the protein–DNA interaction. (B) DNA methylation of the target (ISG15) and off-target (VEGFA) regions analyzed by targeted bis-seq in HEK293 cells using the dCS system with ISG15 sgRNA and DNMT3A wild type and mutants in Ab-3A3L. Results for the wild type DNMT3A are taken from Figure 2D and shown for comparison. Data present the mean of two independent experiments and error bars show the standard deviation. p-Values for comparison of the off-target activity of the mutants with wild type were 9.36 × 10−3 (K766E), 4.19 × 10−4 (R831E), 2.69 × 10−4 (K844E), and 5.05 × 10−4 (R887E) (one-sided t-test assuming unequal variance based on the experimental repetitions). (C) The specificity of the EpiEditors calculated as a ratio of the on- versus off-target DNA methylation based on the data of (B). (D) Off-target methylation analyzed at the VEGFA CGI for Ab-3A3L containing wild type and mutated DNMT3A co-transfected with dCS and the scrambled sgRNA coding vector. See also Figure S2.