Figure 6.
Impaired DNA Looping at TFIIIC-Looped Genes by TFIIIC Depletion or AEs Deletion Abrogates the Reactivation of Gene Expression upon Serum Re-exposure
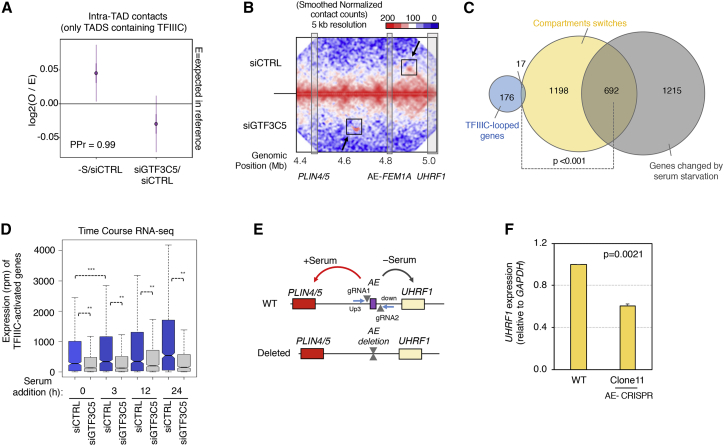
(A) Changes in specific intra-TADs contacts made by TFIIIC in T47D treated with siGTF3C5 or siCTRL in the absence of serum. Data are the log2 FC of observed versus expected (95% CI) of Hi-C data. The changes in siGTF3C5 versus siCTRL show significant (PPr = 0.99) decrease of total intra-TADs contacts compared to mock-transfected (–S) versus siCTRL.
(B) Hi-C matrix heatmaps of the UHRF1 locus for siCTRL and siGTF3C5 cells in the absence of serum. Color scale bar is reported with red representing values from 200 and above, white values equal to 100, and blue values equal to 0. The looping between the AE bound by TFIIIC and its respective targets (PLIN4/5 and UHRF1) is indicated by a box when detected and an arrow when lost. Note that even in the absence of serum the Alu-FEM1A loops back to the PLIN4/5 locus upon siGTF3C5 (as reported for the condition of presence of serum in Figure 4B).
(C) Venn diagram showing the overlap between the TFIIIC-looped and the serum-dependent genes compared to the regions showing compartment switch (Figure S4B). Note the higher overlap of the regions showing compartment changes with the serum-dependent genes (Fisher’s exact test).
(D) RNA-seq expression analysis in T47D cells of TFIIIC-activated genes after serum re-exposure for the indicated times in conditions of siCTRL and siGTF3C5. Expression data are reported as RPM (∗∗p < 1x10−12, two-tailed paired t test in comparisons for each time-point). The comparison between 0 to 3 h in siCTRL cells is shown to highlight the rapidity of gene activation after serum addition (∗∗∗p < 1x10−20). GTF3C5-depleted levels were maintained during the time course (Figure S6B).
(E) Schematic representation of the CRISPR-Cas9 approach to delete the TFIIIC-bound AE located between the PLIN4/5 and UHRF1 loci in chromosome 17. The wild-type (WT) and the deleted alleles are shown. The targeted AE is shown as a purple box, the position of the guide RNAs (gRNA1 and gRNA2) is marked with triangles, and the primers used for the screen (Figure S6C) are indicated with blue arrows. Arrows indicate the chromatin interactions in +S (red) and –S (black) conditions based on Figure 4B.
(F) UHRF1 expression by qRT-PCR in T47D parental cells (WT) and AE-deleted (Clone11) in the absence of serum (mean ± SEM of biological duplicates with data in WT cells normalized to 1; one-tailed t test).