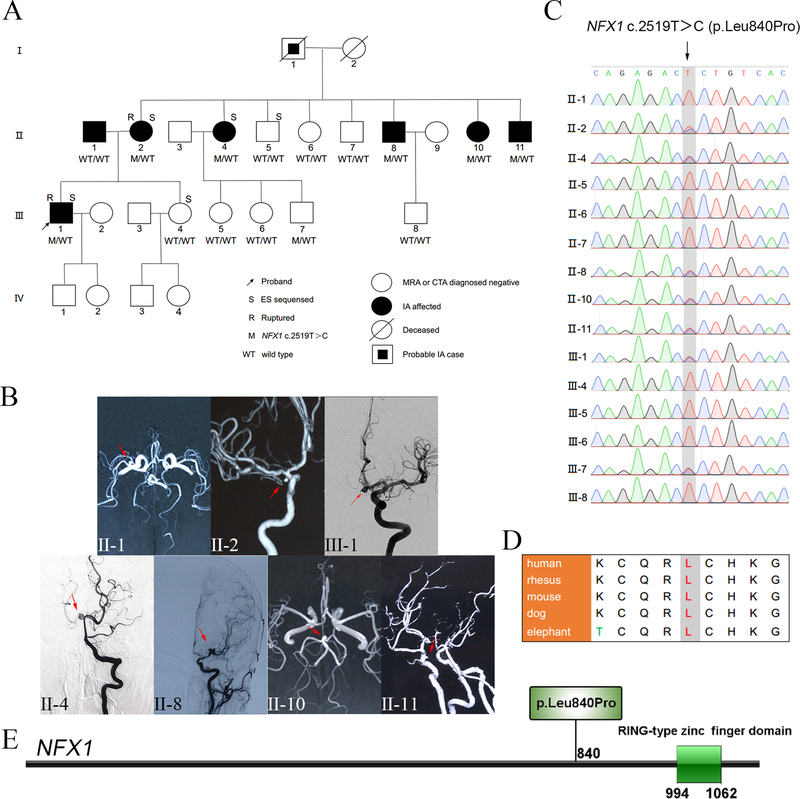
Figure 1.
Family pedigree, clinical images, and segregation results (A-C). Protein location and evolutionary conservation of the NFX1 c.2519T>C variant (D-E). (A) Pedigree and NFX1 genotype segregation in the family. WT/WT represents both wild type alleles, i.e. bi-allelic, while M/WT designates NFX1 c.2519T>C pathogenic variant as heterozygous allele. (B) MRA/DSA images of the presenting family members IA; i.e. clinically affected cases. Red arrows indicate the site of IAs. (C) Validation of the NFX1 c.2519T>C variant via Sanger sequencing. (D) Protein sequence alignments indicate that 840Leu in NFX1 is robustly conserved in mammals. (E) NFX1 p.Leu840Pro mapping, located in a region where no domain structure has been delineated before the RING-type zinc finger domain of NFX1 (http://ibs.biocuckoo.org/).