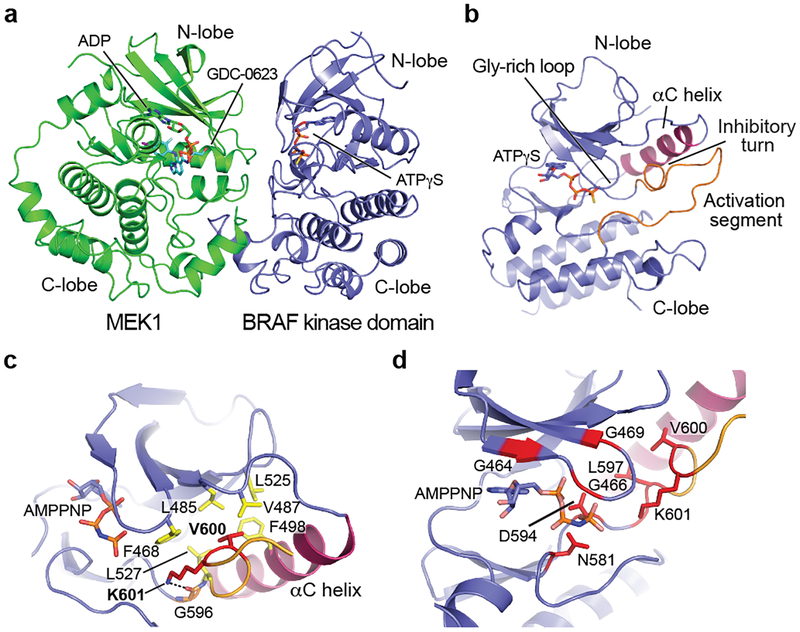
Figure 2. Conformation of the autoinhibited BRAF kinase domain and location of oncogenic mutations.
a, BRAF coordinates MEK1 in a face-to-face orientation, with extensive contact between the kinase C-lobes. Both kinases adopt an inactive, αC-out conformation. b, Overall view of the autoinhibited BRAF kinase domain. The C-helix (purple) is propped in an outward, inactive conformation by the inhibitory turn in the activation segment (orange). ATP-γS is bound in the active site cleft. c, Detailed view of the structure and interactions of the inhibitory turn. Residue V600, the most common site of oncogenic mutations, is part of a cluster of hydrophobic residues (yellow) that stabilize this inactive conformation. K601 also stabilizes this configuration; it hydrogen bonds with G596 in the DFG motif and packs with F468 in the glycine-rich loop. d, Oncogenic mutations (red) cluster in the inhibitory turn or residues that coordinate ATP. Panels a and b are drawn from the cryo-EM structure; panels c and d from the crystal structure of the autoinhibited kinase domain complex.