Figure 5. Neocentromeres are not positioned at sites of preferential CENP-A loading on chromosome arms.
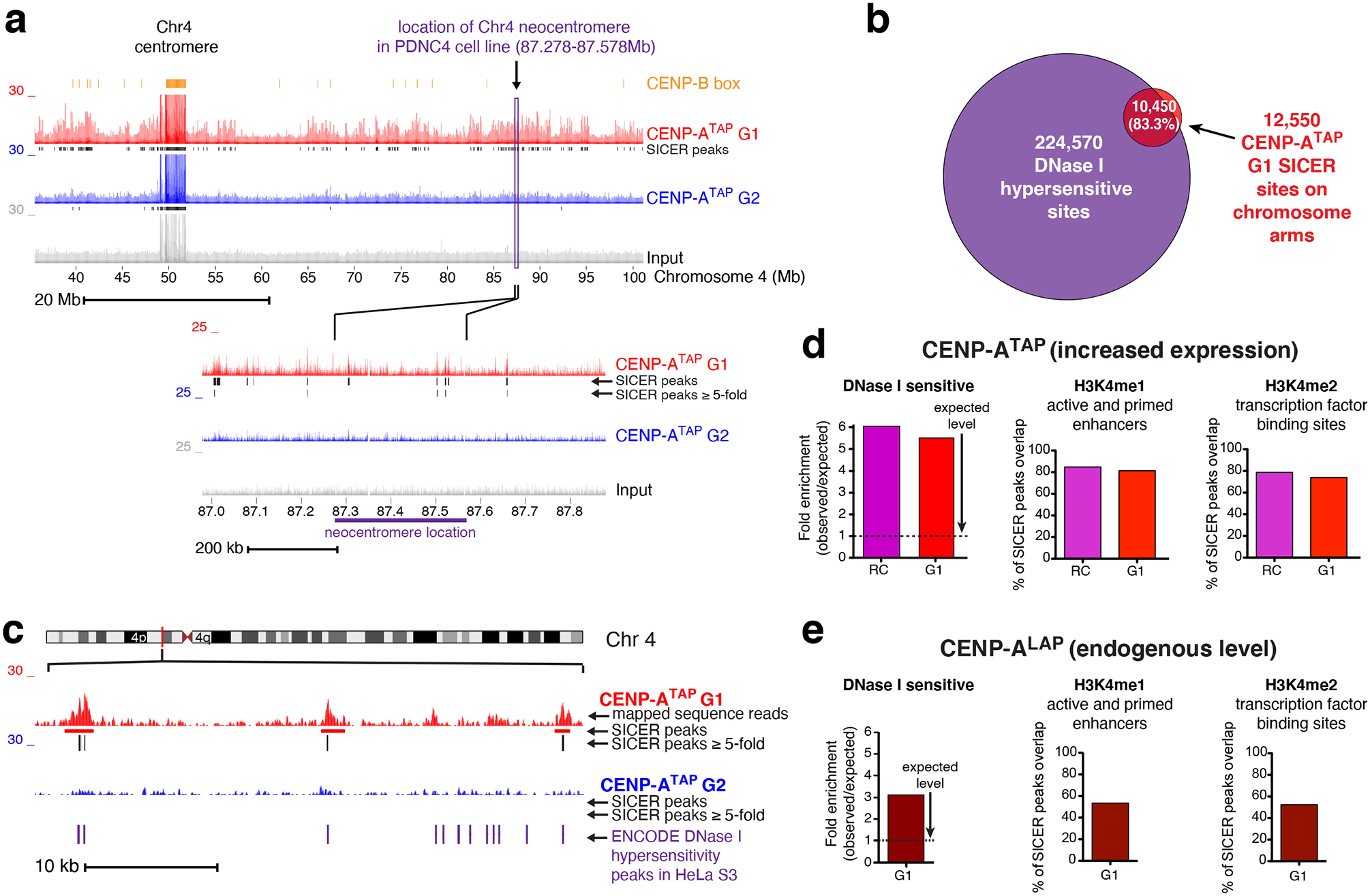
(a) Read mapping data of CENP-ATAP ChIP-sequencing at G1 and G2, at the chromosomal location of a known patient derived neocentromere39 found in chromosome 4. The experiment was repeated independently twice with similar results. (b) Greater than 80% of CENP-A SICER peaks ≥ 5-fold in randomly cycling and G1 cells overlap with DNase I hypersensitive sites taken from ENCODE project. (c) Example from the chromosome 4 p-arm showing overlap of at least 100 bases between SICER peaks ≥ 5-fold and HeLa S3 DNase I hypersensitive sites taken from ENCODE project. The experiment was repeated independently twice with similar results. (d, e) CENP-ATAP (d) or CENP-ALAP (e) enrichment levels at DNase I hypersensitive sites, H3K4me1, and H3K4me2 sites. SICER peaks ≥ 5-fold supported between the two replicates were analyzed for their enrichment level at DNase I hypersensitive sites, with minimum overlap of 100 bases, compared to the level of enrichment at these sites by chance. SICER peaks ≥ 5-fold supported between the two replicates of CENP-ATAP (d) or CENP-ALAP (e) were analyzed for their overlap degree with known sites of H3K4me1 and H3K4me2 binding taken from ENCODE HeLa S3 datasets. Source data for d and e can be found in Supplementary Table 4.
