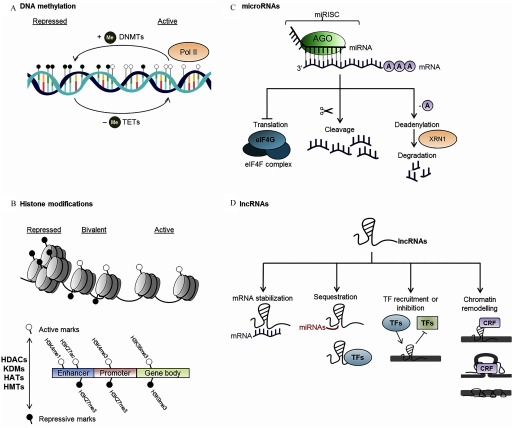
Figure 1.
Diagrammatical summary of epigenetic modes on action. (A) The deposition of methyl groups is mediated by the DNA methyltransferases (DNMTs), and their removal is mediated by the ten-eleven translocation (TET) family proteins (reviewed by Greenberg and Bourc’his 2019; Wu and Zhang 2010). Increased deposition of methyl groups promotes the condensation of chromatin, whereas reduced DNA methylation is associated with increased accessibility to transcription machinery [represented by RNA polymerase II (Pol II)]. DNA hypermethylation generally leads to the silencing of gene expression when it occurs at gene promoters (reviewed by Greenberg and Bourc’his 2019; Wu and Zhang 2010). (B) Histone modifications are deposited by multiple classes of enzymes [histone acetyltransferases (HATs), histone methyltransferases (HMTs), histone deacetylases (HDACs), and histone lysine demethylases (KDMs)] (reviewed by Chen et al. 2017). Examples of histone modifications illustrated here are histone H3 lysine 27 trimethylation (H3K27me3) and H3 lysine 9 trimethylation (H3K9me3), which are usually associated with repression of gene transcription, and H3 lysine 4 monomethylation (H3K4me1), H3 lysine 4 trimethylation (H3K4me3), H3 lysine 36 trimethylation (H3K36me3), and H3 lysine 27 acetylation (H3K27ac) marks that are typically associated with activation of transcription. Simultaneous deposition of opposing histone marks is associated with poised or bivalent chromatin, which is in a transitional state poised to be resolved into active or repressed states (reviewed by Chen et al. 2017). (C) microRNAs (miRNAs) bound to the Argonaute (AGO) protein make up the miRNA-induced silencing complex (miRISC). miRNAs direct the miRISC to target mRNAs base-pairing partially to complementary binding sites, which will be cleaved by catalytically active AGO proteins (O’Brien et al. 2018). Alternatively, AGO proteins can recruit additional protein partners, initiating the process of deadenylation, decapping and 5ʹ-to-3ʹ mRNA degradation by 5ʹ-to-3ʹ exoribonuclease 1 (XRN1) (reviewed by Jonas and Izaurralde 2015). There has also been evidence that miRNAs inhibit translation by inhibiting the eukaryotic initiation factor 4F (eIF4F) complex, although this process has yet to be fully elucidated (reviewed by Jonas and Izaurralde 2015). 5ʹ polyA tails are denoted by circles labeled A. (D) Long noncoding RNAs (lncRNAs) may influence gene expression by increasing or decreasing target mRNA stability, by acting as decoys to miRNA and transcription factors (TFs), thus sequestering them from their cognate promoters, or they may recruit or inhibit TF binding to their target sites on the chromatin (reviewed by Angrand et al. 2015). lncRNAs may also recruit chromatin remodeling factors (CRFs) such as the polycomb repressive complexes or cohesin proteins that recruit histone modifier complexes or initiate long-range chromatin looping (reviewed by Angrand et al. 2015). The Xist lncRNA triggers stable repression of the presumptive inactive X-chromosome by physically coating the X-chromosome (reviewed by Lee and Bartolomei 2013).