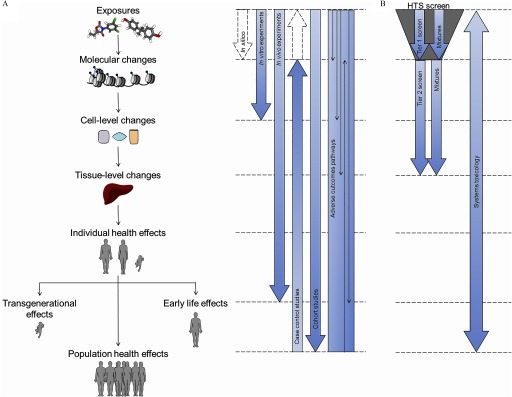
Figure 3.
An integrative method for investigating the impact of environmental chemicals on the epigenome and a proposed approach for toxico-epigenomic screening. (A) Comparison of in silico, in vitro, in vivo, population-based, and integrative methods in fully understanding the potential effects of environmental chemicals on the epigenome. Dotted arrows indicate situations where evidence can be inferred but not directly proved by the described methods. (B) An illustration of a proposed approach for toxicoepigenomic screening. A high-throughput screen (HTS) using in vitro and in silico methods can be conducted using single compounds and mixtures. Hits identified from the Tier 1 screen can be characterized more extensively using relevant in vitro and in vivo experiments. Finally, a systems toxicology approach could be used to integrate all data, including human data, to generate a complete profile of epigenotoxicology.