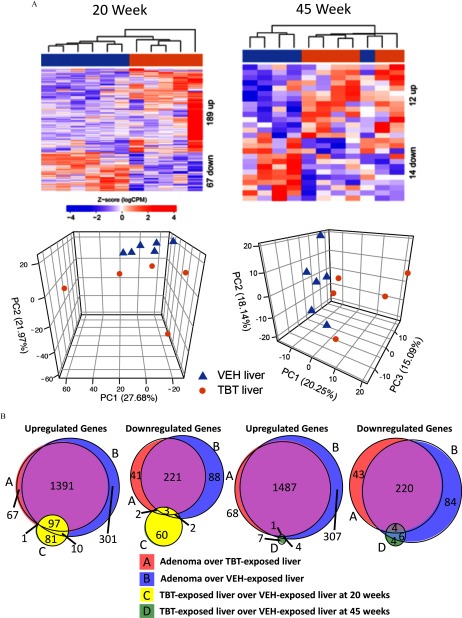
Figure 4.
Differential gene expression in tributyltin (TBT)-exposed liver samples and overlap with adenoma signatures. (A) Upper, hierarchical clustering of RNA-seq data from liver samples from 20- and 45-week-old male mice using differentially expressed genes [false discovery rate (FDR) adjusted with a fold change exceeding ]. Lower, principal component analysis of RNA-seq data at each time point. Triangles represent vehicle (VEH)-exposed liver samples, and circles represent TBT-exposed liver samples. (B) Venn diagrams of overlapping genes in all four gene expression signatures (up-regulated–left, down-regulated–right). A () over TBT-exposed liver (); B () over VEH-exposed liver (); C () over VEH-exposed liver () at 20 weeks; and D () over VEH-exposed liver (). See Excel Table S1 for detail information on n and litter.