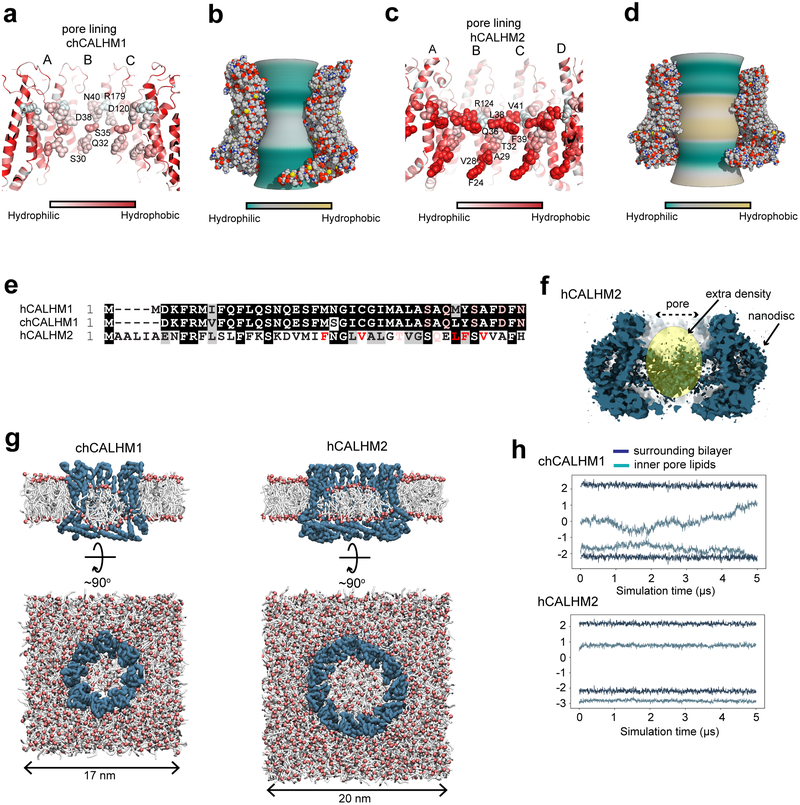
Fig. 5. Comparison of pore properties between chCALHM1 and hCALHM2.
a-d, Channel lining residues (a and c)28 and inner pore surface (b and d; calculated by CHAP29) of chCALHM1 (a-b) and hCALHM2 (c-d) colored based on relative hydrophobicity. These parameters are calculated based on the modeled residues, thus, the amino terminal ends are not taken into account. e, Sequence alignment of the N-terminal residues showing hydrophobic and hydrophilic residues facing the pore with the same color code as in panel a and c. f, Cross-section of the hCALHM2 showing an extra cryo-EM density in the middle of the pore. g, Coarse-grained MD simulations of chCALHM1 (left) and hCALHM2 (right) embedded in POPC membranes. Side (cutaway) and top views of the final frame of one 5 μs replicate are shown in each case, with the protein backbone particles in blue, phospholipid headgroups in red, and acyl tails in white. Water and ions present in the simulation systems are omitted for clarity. h, Headgroup positions of lipids inside each channel pore and in the surrounding bilayer membrane. The average headgroup z-coordinates of lipids constituting the upper- and lower-leaflets in the final frame of each simulation are respectively tracked through the 5 μs simulated duration; results from one replicate are shown for each protein.