Abstract
Increasing evidence has indicated that long non-coding RNAs (lncRNAs) play significant roles in various diseases; however, their roles in age-related macular degeneration (AMD) remain unclear. Dedifferentiation and dysfunction of retinal pigment epithelium (RPE) cells have been shown to contribute to AMD etiology in several studies. Herein, we found that lncRNA LINC00167 was downregulated in RPE-choroid samples of AMD patients and dysfunctional RPE cells, and it was consistently upregulated along with RPE differentiation. In vitro study indicated that reduced endogenous LINC00167 expression resulted in RPE dedifferentiation, which was typified by attenuated expression of RPE markers, reduced vascular endothelial growth factor A secretion, accumulation of mitochondrial reactive oxygen species, and interrupted phagocytic ability. Mechanistically, LINC00167 functioned as a sponge for microRNA miR-203a-3p to restore the expression of the suppressor of cytokine signaling 3 (SOCS3), which further inhibited the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway. Taken together, our study demonstrated that LINC00167 showed a protective role in AMD by maintaining RPE differentiation through the LINC00167/miR-203a-3p/SOCS3 axis and might be a potential therapeutic target for AMD.
Keywords: LINC00167, age-related macular degeneration, retinal pigment epithelium, miR-203a-3p, SOCS3
Introduction
Retinal pigment epithelium (RPE) is composed of cuboidal, postmitotic, and polarized cells located in the outer retina between photoreceptor outer segments and Bruch’s membrane.1, 2, 3 RPE plays vital roles in maintaining balance of the retinal microenvironment through phagocytosis and digestion of photoreceptor outer segments, participating in the visual cycle, composing the blood-retina barrier with tight junctions, and secreting multiple growth factors essential for endothelial cells.1,4 Depletion of RPE function is involved in multiple retinal degenerative diseases, including age-related macular degeneration (AMD).1,5, 6, 7 AMD is one of the leading causes for vision loss in aged people.8 Due to its irreversible progression and unsatisfied treatment effect, AMD severely lowered patients’ life quality.9,10 According to histopathologic changes in the retina, AMD can be classified in to two categories: nonexudative AMD and neovascular AMD.11 Nonexudative AMD, also known as dry AMD, is characterized by drusen or debris within Bruch’s membrane, subretinal deposits, RPE abnormalities, and RPE cell loss.12, 13, 14 By far, no efficient treatment has been raised for dry AMD. Previous study has illuminated that RPE dedifferentiation marked by attenuated RPE-characteristic protein is associated with RPE dysfunction in the early stage of AMD.1,15 Therefore, investigations into RPE dedifferentiation may help to learn more about AMD pathogenesis and to come up with effective therapy for AMD.
Long non-coding RNAs (lncRNAs) are a class of non-protein-coding transcripts longer than 200 nt.16 Functions of lncRNAs include co-transcriptional regulation, molecular recruitment to specific loci, RNA-binding titration, and microRNA (miRNA) sponge.16,17 lncRNAs could regulate messenger RNA (mRNA) expressions by functioning as a miRNA sponge,18,19 and they play regulatory roles in various biological processes, such as tumor genesis, cell differentiation, and epithelial-mesenchymal transition.20, 21, 22 However, roles of lncRNAs in RPE dedifferentiation remain unclear. In this study, we sought to investigate the function of lncRNA LINC00167 in RPE dedifferentiation.
Results
LINC00167 Is Decreased in AMD
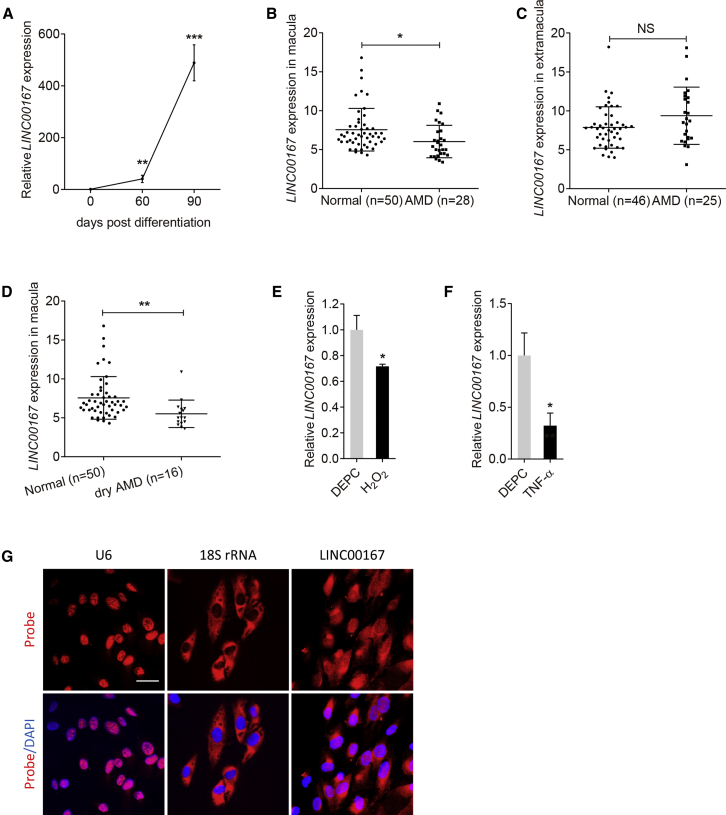
RPE dedifferentiation is an initial pathological event in AMD.1,3 We have previously identified the crucial roles of lncRNAs in AMD etiology.14 Herein, we analyzed the expression of lncRNA LINC00167 (ENST00000530583) in human-induced pluripotent stem cells (hiPSCs) and in hiPSC-induced RPE cells at 30, 60, and 90 days post-differentiation, respectively. We found that LINC00167 expression was consistently upregulated along with RPE differentiation (Figure 1A). To better illustrate its role in AMD pathogenesis, we further compared its expression levels in RPE-choroid samples between AMD patients and normal controls. Clinical diagnosis and personal information of all individuals are presented in Table S1.23 We found that LINC00167 expression was downregulated in the macular RPE-choroid tissue of AMD patients compared to normal controls (Figure 1B), but not in the extramacular tissue (Figure 1C). Our data also revealed that expression of LINC00167 was mainly reduced in the macular RPE-choroid tissue of dry AMD (Figure 1D). We next measured its expression pattern in dysfunctional RPE. We found that LINC00167 expression was decreased in H2O2 or tumor necrosis factor α (TNF-α)-treated RPE cells (Figures 1E and 1F). Thus, our results suggested a potentially protective role of LINC00167 in AMD pathogenesis.
Figure 1.
LINC00167 Is Decreased in AMD
(A) Expression of LINC00167 along with RPE differentiation. (B) LINC00167 expression in macular RPE-choroid tissues of normal controls and AMD patients. (C) LINC00167 expression in extramacular RPE-choroid tissues of normal controls and AMD patients. (D) LINC00167 expression in macular RPE-choroid tissues of normal controls and dry AMD patients. (E) Expression of LINC00167 in diethyl pyrocarbonate (DEPC)- and H2O2-treated RPE cells. (F) Expression of LINC00167 in DEPC- and TNF-α-treated RPE cells. (G) Intracellular localization of U6, 18S rRNA, and LINC00167 in RPE cells as indicated by RNA-FISH. Scale bar, 20 μm. The data are presented at as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. NS, not significant.
Function of an lncRNA mainly depends on its subcellular location. lncRNAs located in nucleus may act as neighbor gene modulators through promoter interactions, and lncRNAs expressed in cytoplasm may function as miRNA sponges.17 According to fluorescence in situ hybridization (FISH) results, LINC00167 was mainly located in cytoplasm (Figure 1G), indicating its potential function as a sponge for miRNA.
LINC00167 Silencing Leads to RPE Dedifferentiation
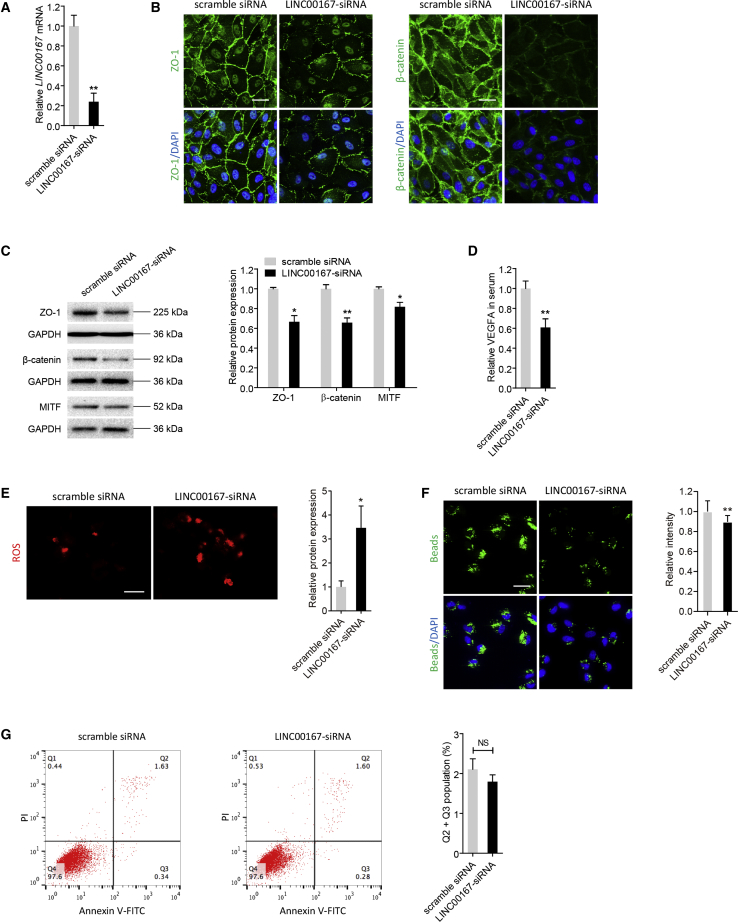
We next tried to determine the effects of LINC00167 on RPE differentiation. Quantitative real-time PCR showed a 75% reduction of LINC00167 expression in adult RPE-19 (ARPE-19) cells transfected with LINC00167-small interfering RNA (siRNA) compared to cells transfected with scramble siRNA (Figure 2A). We then adopted immunoblotting and immunofluorescence to compare expressions of RPE characteristic markers, including tight junction protein ZO-1 (NP_003248), β-catenin (NP_001895), and microphthalmia-associated transcription factor (MITF; NP_001341533), between the LINC00167-siRNA-transfected group and the scramble siRNA-transfected group. Based on our data, endogenous LINC00167 insufficiency suppressed expressions of those markers (Figures 2B and 2C). Our findings suggested that LINC00167 promoted differentiation of RPE cells.
Figure 2.
LINC00167 Silencing Leads to RPE Dedifferentiation
(A) Relative expression of LINC00167 in ARPE-19 cells transfected with LINC00167-siRNA compared to cells transfected with scramble siRNA. (B) Expressions and intracellular localizations of RPE markers ZO-1 and β-catenin were compared between ARPE-19 cells transfected with LINC00167-siRNA and scramble RNA using immunofluorescence staining. Scale bars, 20 μm. (C) Immunoblotting was applied to compare expression levels of ZO-1, β-catenin, and MITF between ARPE-19 cells transfected with LINC00167-siRNA and scramble RNA. A representative image and the quantification results are shown. (D) Secreted VEGFA levels in serum of ARPE-19 cells transfected with LINC00167-siRNA and scramble siRNA. (E) Mitochondrial ROS was visualized in ARPE-19 cells transfected with LINC00167-siRNA and scramble siRNA. A representative image and the quantification results are shown. Scale bar, 50 μm. (F) Phagocytic ability was tested in ARPE-19 transfected with LINC00167-siRNA and scramble siRNA. A representative image and the quantification results are shown. Scale bar, 20 μm. (G) Apoptosis rates were monitored by flow cytometric analysis in ARPE-19 cells transfected with LINC00167-siRNA and scramble siRNA. A representative image and the quantification results are shown. The data are presented at as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01. NS, not significant.
We next tested whether LINC00167 insufficiency would cause other forms of RPE abnormalities. Secretion of vascular endothelial growth factor A (VEGFA) is an essential function of RPE cells,4 which maintains the health of choriocapillaris endothelium. Insufficient VEGFA secretion is an important contributing factor for dry AMD. We therefore used an enzyme linked immunosorbent assay (ELISA) to determine VEGFA secretion of RPE cells in culture medium. A decreased amount of VEGFA was found in RPE cells with LINC00167 knocked down compared to the control group (Figure 2D). Oxidative stress, which leads to accumulation of mitochondrial reactive oxygen species (ROS), contributes to RPE dysfunction and AMD pathogenesis.1,5 Herein, we found that ROS generation was increased in RPE cells with LINC00167 silenced (Figure 2E). Another crucial function of RPE cells is phagocytizing photoreceptor outer segment debris, which maintains retinal homeostasis. Impaired RPE phagocytosis leads to deposition of apolipoprotein B100 and formation of drusen and basal deposits, which are important histopathologic changes in dry AMD.24,25 According to our results, attenuated phagocytosis was revealed in RPE cells with endogenous LINC00167 insufficiency when compared to cells transfected with scramble siRNA (Figure 2F). To rule out the possibility that such disturbed phagocytosis was caused by RPE cell death, we next measured RPE apoptosis rates in different transfected groups. No statistical difference in apoptosis rates was detected between RPE cells transfected with LINC00167-siRNA and scramble siRNA (Figure 2G). Thus, LINC00167 insufficiency impaired phagocytosis independent of RPE cell death.
LINC00167 Functions as a Sponge for miR-203a-3p in RPE Cells
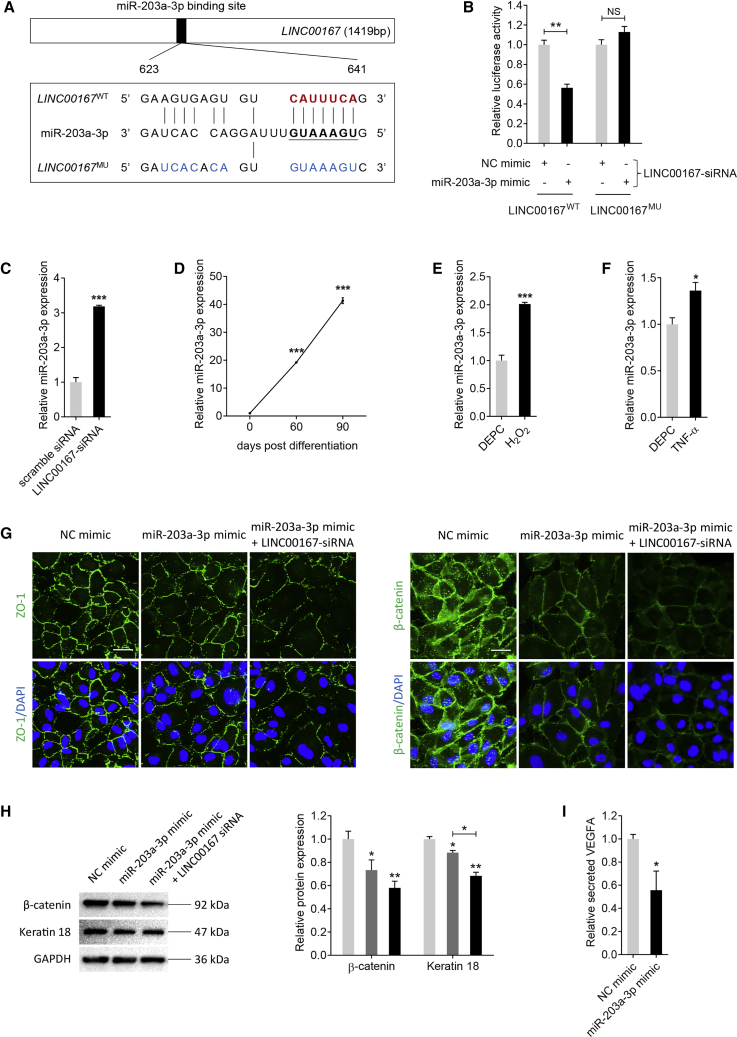
lncRNAs with miRNA-binding sites might function as miRNA sponges26 and are involved in lots of biological processes and disease etiologies.27 As LINC00167 was mainly localized in the cytoplasm, we hypothesized that it might act as a miRNA sponge in RPE cells. miR-203a-3p was revealed as a potential target of LINC00167 as predicted by miRcode online software (http://mircode.org/). We initially verified the interaction between LINC00167 and miR-203a-3p using luciferase reporter assay. LINC00167MU plasmid contained 13 mutated nucleotides in the core binding region with miR-203a-3p (Figure 3A). According to our data, luciferase activity was reduced in RPE cells co-transfected with wild-type LINC00167 (LINC00167WT), miR-203a-3p mimic, and LINC00167-siRNA compared to cells co-transfected with LINC00167WT, negative control (NC) mimic, and LINC00167-siRNA (Figure 3B). Introduction of the mutation abolished the ability of LINC00167 to bind to miR-203a-3p (Figure 3B). We then tried to determine whether LINC00167 expression was negatively correlated with miR-203a-3p. We found that miR-203a-3p was upregulated in cells transfected with LINC00167 siRNA compared to scramble siRNA (Figure 3C). We also found that miR-203a-3p was consistently upregulated along with RPE differentiation (Figure 3D), and was increased in RPE cells treated with H2O2 or TNF-α (Figures 3E and 3F). Taken together, our data suggested that LINC00167 acted as a miR-203a-3p sponge in RPE cells.
Figure 3.
LINC00167 Functions as a Sponge for miR-203a-3p in RPE Cells
(A) Schematic diagram of the interaction between LINC00167 and miR-203a-3p. (B) Relative luciferase activities in ARPE-19 cells transfected with LINC00167-siRNA and LINC00167WT or LINC00167MU and miR-203a-3p mimic or NC mimic. (C) Relative miR-203a-3p expression in cells transfected with LINC00167-siRNA and scramble RNA. (D) Expression of miR-203a-3p along with RPE differentiation. (E) Expression of LINC00167 in diethyl pyrocarbonate (DEPC)- and H2O2-treated RPE cells. (F) Expression of LINC00167 in DEPC- and TNF-α-treated RPE cells. (G) RPE characteristic markers ZO-1 and β-catenin were observed by immunofluorescence staining in ARPE-19 cells transfected with NC mimic, miR-203a-3p mimic, and miR-203a-3p mimic together with LINC00167-siRNA. Scale bars, 20 μm. (H) Expression levels of RPE markers β-catenin and keratin 18 were monitored by immunoblotting in ARPE-19 cells transfected with NC mimic, miR-203a-3p mimic, and miR-203a-3p mimic together with LINC00167-siRNA. (I) Secreted VEGFA levels in serum of ARPE-19 cells transfected with NC mimic and miR-203a-3p mimic. The data are presented at as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. NS, not significant.
The LINC00167/miR-203a-3p/SOCS3 Axis Regulates RPE Differentiation
We then analyzed the role of miR-203a-3p on RPE differentiation. According to our data, exogenous miR-203a-3p overexpression led to downregulation of RPE characteristic proteins, including ZO-1, β-catenin, and keratin 18, as revealed by immunofluorescence and immunoblotting (Figures 3G and 3H). We also found that silencing of LINC00167 and overexpression of miR-203a-3p at the same time could show double hit effects on RPE cells. Expression of RPE markers was more reduced in RPE cells transfected with both LINC00167 siRNA and miR-203a-3p mimic (Figures 3G and 3H). Our data also revealed that VEGFA secretion was disturbed in RPE cells transfected with miR-203a-3p mimic when compared to the control group (Figure 3I).
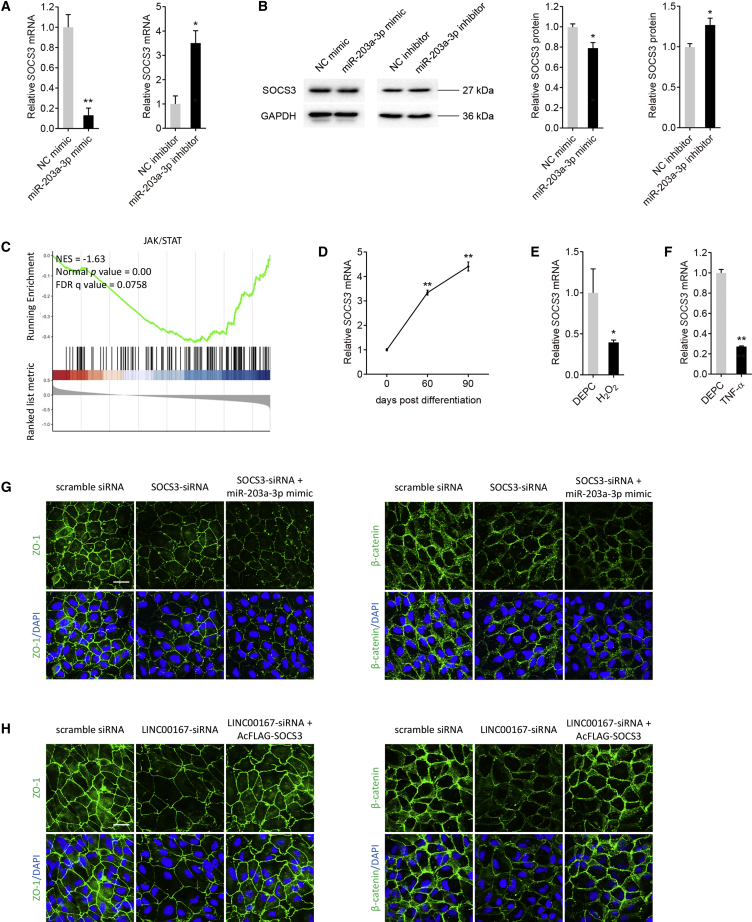
The suppressor of cytokine signaling 3 (SOCS3) gene was previously identified as a direct target of miR-203a-3p.28,29 SOCS3 was reported to show a protective role in AMD etiology by regulating the Janus kinase/signal transducer and activator of transcription (JAK/STAT) cell signaling pathway.30 We therefore tested whether SOCS3 was a target of miR-203a-3p in RPE cells. Expressions of both SOCS3 mRNA and protein were decreased in RPE cells transfected with miR-203a-3p mimic and increased in RPE cells transfected with miR-203a-3p inhibitor (Figures 4A and 4B), showing a reverse expressional correlation between SOCS3 and miR-203a-3p. Our data suggested that SOCS3 was negatively regulated by miR-203a-3p.
Figure 4.
miR-203a-3p/SOCS3 Axis Regulates RPE Differentiation
(A and B) Relative expression of SOCS3 mRNA (A) and protein (B) in ARPE-19 cells transfected with NC mimic, miR-203a-3p mimic, NC inhibitor, and miR-203a-3p inhibitor. (C) JAK/STAT enrichment analysis. (D) Expression of SOCS3 along with RPE differentiation. (E) Expression of SOCS3 in diethyl pyrocarbonate (DEPC)- and H2O2-treated RPE cells. (F) Expression of SOCS3 in DEPC- and TNF-α-treated RPE cells. (G) We used immunofluorescence staining to visualize RPE characteristic markers ZO-1 and β-catenin in ARPE-19 cells transfected with scramble siRNA, SOCS3-siRNA, and SOCS3-siRNA together with miR-203a-3p mimic. Scale bar, 20 μm. (H) Immunofluorescence staining was applied to compare expressions of ZO-1 and β-catenin in ARPE-19 cells transfected with scramble siRNA, LINC00167-siRNA, and LINC00167-siRNA together with AcFLAG-SOCS3. Scale bar, 20 μm. The data are presented at as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
We also used gene set enrichment analysis (GSEA) to speculate on possible signaling pathways associated with LINC00167. The JAK/STAT pathway, in which SOCS3 is a key negative regulator, has a negative correlation with LINC00167 (Figure 4C). Taking all off the above findings into consideration, SOCS3 was identified as a downstream part of LINC00167/miR-203a-3p.
We next investigated its role in mediating RPE function. SOCS3 mRNA expression was consistently upregulated along with RPE differentiation (Figure 4D), and it was downregulated in H2O2- or TNF-α-treated RPE cells (Figures 4E and 4F). SOCS3 siRNA was applied to reduce endogenous SOCS3 expression in RPE cells (Figure S1A), and the AcFLAG-SOCS3 vector was used to overexpress SOCS3 in RPE cells (Figure S1B). According to our results, RPE characteristic proteins, including ZO-1 and β-catenin, were degraded in RPE cells transfected with SOCS3 siRNA (Figure 4G). We further aimed to tell whether miR-203a-3p regulated RPE function through SOCS3. Protein levels of ZO-1 and β-catenin were more reduced in RPE cells co-transfected with SOCS3 siRNA and miR-203a-3p when compared to cells transfected with SOCS3 siRNA alone (Figure 4G). To better tell whether SOCS3 is the downstream target of LINC00167, we tested whether overexpression of SOCS3 could rescue the LINC00167 knockdown phenotype. According to our data, overexpression of SOCS3 could rescue the negative effect of LINC00167-siRNA on RPE cells (Figure 4H). Thus, our findings indicated that the LINC00167/miR-203a-3p/SOCS3 axis participated in RPE differentiation.
Discussion
RPE dedifferentiation is revealed as an initial pathological event for AMD.1 Therefore, illustration of the mechanism underlying RPE dedifferentiation may help with the identification of therapeutic targets for AMD. lncRNAs are involved in the pathogenesis of multiple diseases, including carcinoma, neural degeneration, and cardiovascular and ocular diseases,16,17,21,31 while its role in AMD etiology and RPE dedifferentiation has not been fully elucidated. We have previously found that lncRNA ZNF503-AS1 could promote RPE differentiation by downregulating ZNF503 expression.14 However, more investigations in this area are still warranted. In this study, we revealed that LINC00167 was downregulated during RPE differentiation and mediated RPE differentiation through the LINC00167/miR-203a-3p/SOCS3 axis.
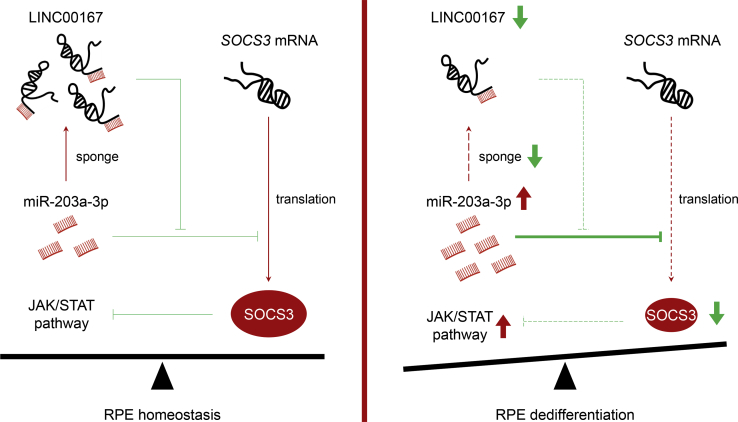
LINC00167 belongs to intergenic lncRNAs that possess an exon-intron-exon structure but do not code for proteins.32 In the present study, we found that LINC00167 expression was constantly upregulated along with RPE differentiation, and it was decreased in RPE-choroid samples of AMD patients and in H2O2- or TNF-α-treated RPE cells. In vitro study revealed that LINC00167 insufficiency led to RPE dedifferentiation, which attenuated RPE function by impairing phagocytosis, disturbing VEGFA secretion, and accelerating ROS production. As LINC00167 was found mainly localized in RPE cytoplasm, we speculated that it might function as a competitive endogenous RNA (ceRNA). ceRNAs are lncRNAs that share one or more miRNA response elements with mRNAs and function as natural pseudogenes to regulate miRNA activities.18,31 Involvement of lncRNA-miRNA-mRNA loops in ocular diseases have been previously revealed.33,34 In this study, we first revealed that LINC00167 inhibited RPE dedifferentiation and AMD progression by sponging miR-203a-3p to block its interaction with SOCS3 (Figure 5).
Figure 5.
Schematic Illustration of the LINC00167/miR-203a-3p/SOCS3 Axis in Normal RPE Cells and Dedifferentiated RPE Cells
miR-203a-3p binds to both LINC00167 and the 3′ UTR of SOCS3. When expression of LINC00167 is downregulated, an increased amount of miR-203a-3p would bind to the SOCS3 3′ UTR to decrease SOCS3 protein expression in a post-transcriptional way. This would lead to RPE dedifferentiation.
miRNAs are small non-coding RNAs that regulate protein production through binding to the 3′ untranslated region (3′ UTR) of mRNA.35 miR-203a-3p has been shown to participate in diverse diseases, including cancer, ocular diseases, and cardiovascular diseases.29,36, 37, 38 Herein, we first revealed that overexpression of miR-203a-3p resulted in RPE dedifferentiation. A previous study revealed that miR-203a-3p was decreased in an oxygen-induced retinopathy model and mechanically repressed angiogenesis by targeting VEGFA and hypoxia-inducible factor-1α (HIF-1α).37 miR-203a-3p was identified to specifically bind to the 3′ UTR of VEGFA in human retinal microvascular endothelial cells. Similar to these findings, we also found that miR-203a-3p overexpression in RPE cells attenuated VEGFA secretion.
AMD is a comprehensive disease with both genetic and non-genetic components involved in its pathogenesis.39,40 Other than epigenetic regulations, other factors, such as genetic variants and environmental variables, have also been found to be associated with AMD susceptibility. Of note, polymorphisms within precursor (pre)-miRNA sequences have been reported to contribute to AMD etiology.41,42 Such polymorphisms might affect the functional activities of miRNAs, especially the interaction of miRNAs with specific targets. Multivariate risk indexes might cooperate together to alter signaling pathways such as complement activation, inflammation, VEGFA signaling, and oxidative stress,40 and further contribute to the pathogenesis of AMD. The pharmacogenetic relationship between genetic variants and the variable response of AMD patients to anti-VEGF therapies has been widely investigated.43 Due to the regulatory role of miR-203a-3p on VEGFA expression, genetic variants in the LINC00167/miR-203a-3p axis might also function as a pharmacoepigenetic biomarker for AMD patients to determine the dosage and administration frequency of anti-VEGF drugs.
We also identified SOCS3 as a direct target of miR-203a-3p in RPE cells. SOCS3 has been revealed as an inhibitor of the JAK/STAT pathway in various studies.30,44, 45, 46, 47, 48, 49, 50, 51, 52 SOCS3 was previously reported to show a protective effect on AMD pathogenesis by regulating the JAK/STAT pathway.30 A recent study also revealed that SOCS3 deficiency promoted inflammation-related retinal degeneration in experimental autoimmune uveoretinitis mice.53 In mammals, the JAK/STAT signaling pathway is reported to participate in various biological processes, including inflammation, cell differentiation, proliferation, migration, and apoptosis.54 Previous studies have identified an inhibitory role of the JAK/STAT pathway in cell differentiation. Activation of the JAK/STAT pathway reversed differentiated spermatogonia into stem cell-like status.48 Partial inhibition of the JAK/STAT pathway by SOCS3 disturbed dedifferentiation of spermatogonia into germline stem cells in Drosophila testis.51 Herein, our study demonstrated that oxidative stress and inflammation downregulated SOCS3 expression in RPE cells. We also revealed that SOCS3 was regulated by the LINC00167/miR-203a-3p axis and mediated RPE dedifferentiation by repressing the JAK/STAT pathway. Therefore, JAK/STAT inhibitors show promising prospects in AMD treatment. However, LINC00167 might have other sponging miRNAs besides miR-203a-3p, and the LINC00167/miR-203a-3p axis might regulate RPE function through other pathways in addition to the SOCS3/JAK/STAT pathway. Thus, more investigations are warranted to better elucidate the regulatory network of LINC00167 in RPE cells. Another limitation of this study is the lack of patient samples to verify the downstream regulatory network of LINC00167 in patients. We will keep working on that in our future studies.
In conclusion, our study revealed that LINC00167 inhibited RPE dedifferentiation and AMD progression by sponging miR-203a-3p to block its interaction with SOCS3. Thus, LINC00167 might be a potential therapeutic target for AMD.
Materials and Methods
Samples
Sample information and microarray data (GEO: GSE29801) of 78 independent macular and 71 extramacular RPE-choroid samples were downloaded from Gene Expression Omnibus datasets and analyzed as described previously.23 Information of all samples and their LINC00167 expression levels are detailed in Table S1.
Cell Culture, Treatment, and Transfection
hiPSCs (IMR90-57) were grown on mouse embryonic fibroblasts (SiDan-Sai Biotechnology, Shanghai, China) in a six-well plate as described before.55 In vitro differentiation of hiPSCs to RPE cells was conducted using the SFEB/CS method. CKI-7 (5 μM) and SB-431542 (5 μM) were added to trigger differentiation. ARPE-19 cells purchased from American Type Culture Collection were cultured in Dulbecco’s modified Eagle’s/F12 medium supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and 1% penicillin/streptomycin. Cells were incubated at 37°C in 5% CO2. For the H2O2 assay, ARPE-19 cells were incubated with H2O2 at the concentration of 200 μM for 48 h before collection. For the TNF-α assay, cells were treated with TNF-α at the concentration of 100 ng/mL for 48 h before collection. For the transfection assay, cells were seeded into six-well templates and transfected with 100 pmol of siRNA/mimic/inhibitor and/or 4 μg of plasmid, using Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA) per well according to the manufacturer’s protocol. NC mimic and inhibitor, miR-203a-3p mimic and inhibitor (2′-O-methyl modification), scramble siRNA, LINC00167-siRNA, and SOCS3-siRNA were purchased from RiboBio (Guangzhou, China) with sequences detailed in Table S2. The open reading frame sequence of SOCS3 was synthesized and inserted into pCMV-C-FLAG plasmid (Beyotime, Shanghai, China) to produce the recombinant plasmid AcFLAG-SOCS3. Cells were collected at 48 h post-transfection for RNA extraction, and at 72 h post-transfection for immunoblotting, immunofluorescence staining, apoptosis analysis, ROS measurement, and luciferase reporter assay.
RNA Extraction, Reverse Transcriptase PCR (RT-PCR), and Quantitative Real-Time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. A NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used to detect concentration and quality of RNA samples. 1 μg of total RNA was used for RT-PCR using a PrimeScript RT kit (Takara, Otsu, Shiga, Japan). Quantitative real-time PCR was conducted using FastStart Universal SYBR Green Master (Rox; Roche, Basel, Switzerland) with the StepOne Plus real-time PCR system (Applied Biosystems, Darmstadt, Germany). Primers used in this study were LINC00167 (forward, 5′-TCAGCTCACTCCTTAACCGC-3′; reverse, 5′-TCTCTCTGCCATCTAGCTGC-3′), 18S ribosomal RNA (18S rRNA; forward, 5′-TTAATTCCGATAACGAACGAGA-3′; reverse, 5′-CGCTGAGCCAGTCAGTGTAG-3′), SOCS3 (forward, 5′-CACTCTCCAGCATCTCTGTC-3′; reverse, 5′-TCGTACTGGTCCAGGAACTC-3′), GAPDH (forward, 5′-CAGCCTCAAGATCATCAGCA-3′; reverse, 5′-TGTGGTCATGAGTCCTTCCA-3′). Bulge-loop miRNA RT-PCR primer sets (one RT primer and a pair of quantitative real-time PCR primers for each set) were designed by RiboBio.
FISH
LINC00167, U6, and 18S rRNA FISH probes were purchased from RiboBio. FISH was conducted according to the manufacturer’s protocol (RiboBio). ARPE-19 cells were seeded into a 12-well plate, fixed with 4% paraformaldehyde (PFA), permeabilized in 0.5% Triton X-100 for 10 min on ice, and then covered with pre-hybridization buffer. After removal of pre-hybridization buffer, cells were hybridized with a Cy3-labeled LINC00167 probe and placed in a 37°C incubator overnight. Cell nuclei were counterstained by 4′,6-diamidino-2-phenylindole (DAPI). Images were taken with a confocal microscope (LSM 510; Carl Zeiss, Jena, Germany).
Immunofluorescence Staining
Cells were fixed with 4% PFA, permeabilized with 0.5% Triton X-100, blocked in 5% bovine serum albumin, and incubated in primary antibody reagents at 4°C overnight. Details for antibodies are provided in Table S3. Cells were then washed with phosphate-buffered saline (PBS) three times and incubated in corresponding fluorescence-conjugated secondary antibodies (1:1,000 diluted in 1× PBS; Invitrogen) for 1 h at room temperature. Cell nuclei were stained by DAPI (Sigma, St. Louis, MO, USA). Images were collected by a confocal microscope (LSM 510).
Immunoblotting
Extracted protein was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Membranes were blocked in 5% skim milk at 37°C for 1 h and incubated with primary antibody reagents at 4°C overnight. Details for antibodies are provided in Table S3. After incubation, membranes were washed with Tris-buffered saline with Tween 20 (TBST) and probed with corresponding horseradish peroxidase-conjugated secondary antibodies (1:5,000 diluted in TBST; ICL, Newberg, OR, USA) for 2 h at room temperature. Blots were then developed by autoradiography with the enhanced chemiluminescence (ECL)-western blotting system (Bio-Rad, Hercules, CA, USA) following the manufacturer’s protocol. ImageJ software (https://imagej.nih.gov/ij/index.html) was utilized to measure protein intensities.
Mitochondrial ROS Measurement
ARPE-19 cells were grown on 12-well plates. Mitochondrial ROS were detected using the MitoSOX Red mitochondrial superoxide indicator (Invitrogen) according to the manufacturer’s protocol. Cells were treated with 5 μM MitoSOX probe at 37°C for 10 min. A Leica DM4000 B LED microscope (Leica, Wetzlar, Germany) was used to detect ROS signal.
Analyses of Phagocytosis
Phagocytic ability was conducted according to a previously described protocol.3 Briefly, ARPE-19 cells were planted on eight-well chamber slides (Millipore, Billerica, MA, USA) and harvested at 72 h post-transfection. Cells were then incubated with carboxylate-modified polystyrene latex beads (diameter, 1 μm; emission maximum, 515 nm; Sigma) at 37°C for 12 h, washed with 1× PBS, and then treated with 0.2% trypan blue to quench extracellular fluorescence. Cell nuclei were counterstained with DAPI. An LSM 510 confocal microscope was applied for image collection. We used ImageJ software to quantify fluorescence.
Apoptosis Analysis
ARPE-19 cells were harvested and incubated with annexin V-fluorescein isothiocyanate (FITC; Vazyme, Nanjing, China) and propidium iodide (Vazyme) according to the manufacturer’s protocol. Flow cytometric analysis was then performed to examine apoptotic cells using a Gallios flow cytometry (Beckman Coulter, Brea, CA, USA). A total of 10,000 living cells were collected for examination. Data were plotted and analyzed using FlowJo v10 software. Annexin V-FITC-positive cells were considered as apoptotic cells.
Luciferase Reporter Assay
The luciferase reporter assay was conducted according to a previously described protocol.14 The entire LINC00167 sequence was synthesized and inserted into the pGL3-promoter vector (Promega, Madison, WI, USA) using the XbaI restriction site to generate the recombinant plasmids LINC00167WT and LINC00167MU. The LINC00167MU contained seven mutated nucleotides in the core binding region with miR-203a-3p. Constructed plasmids were sequenced and confirmed with Sanger sequencing. ARPE-19 cells were seeded into 24-well plates and transfected with 16 ng of cytomegalovirus-Renilla (Promega), 20 pmol of miR-203a-3p mimic or NC mimic, and 800 ng of LINC00167WT or LINC00167MU per well using Lipofectamine 3000 transfection reagent (Invitrogen). Luciferase activities were measured by a dual-luciferase system (Promega) using a GloMax-96 luminometer. Renilla luciferase activities were used as internal standard indicators for transfection efficiency. Firefly luciferase activities were then normalized to Renilla luciferase activities.
ELISA
ARPE-19 cells were grown on a six-well plate with culture medium and were collected at 72 h post-transfection for ELISA. The expression level of VEGFA was determined using a commercial human VEGFA ELISA kit (Beijing 4A Biotech, Beijing, China) according to the manufacturer’s protocol.
Bioinformatics Analysis
GSEA (http://software.broadinstitute.org/gsea/msigdb/index.jsp) was used to identify enriched gene sets in the Kyoto Encyclopedia of Genes and Genomes based on the Pearson’s correlation coefficient with LINC00167 and expression profile of mRNAs in previous study was used as input.23 GraphPad Prism (version 8.0; GraphPad, San Diego, CA, USA) was used for statistical analysis. One-way analysis of variance and a two-tailed Student’s t test were used for comparisons between different groups. All experiments were performed in triplicate, with data being averaged. A p value <0.05 was considered as statistically significant.
Author Contributions
X.C. and Q.L. conceived and designed the study. X.C. and R.S. conducted experiments, analyzed data, and drafted the manuscript. Q.L. interpreted the data and revised the manuscript. D.Y. and C.J. coordinated and analyzed the data. All the authors contributed to, read, and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81770973 to Q.L and 81700877 to X.C); the National Key Research and Development Program of China (2017YFA0104100 to Q.L); the Natural Science Foundation of Jiangsu Province (BK20171087 to X.C); the Six Talent Peaks Project in Jiangsu Province (WSW-004 to X.C); and a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.12.040.
Supplemental Information
References
- 1.Zhao C., Yasumura D., Li X., Matthes M., Lloyd M., Nielsen G., Ahern K., Snyder M., Bok D., Dunaief J.L. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 3.Jiang C., Xie P., Sun R., Sun X., Liu G., Ding S., Zhu M., Yan B., Liu Q., Chen X., Zhao C. c-Jun-mediated microRNA-302d-3p induces RPE dedifferentiation by targeting p21Waf1/Cip1. Cell Death Dis. 2018;9:451. doi: 10.1038/s41419-018-0481-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saint-Geniez M., Kurihara T., Sekiyama E., Maldonado A.E., D’Amore P.A. An essential role for RPE-derived soluble VEGF in the maintenance of the choriocapillaris. Proc. Natl. Acad. Sci. USA. 2009;106:18751–18756. doi: 10.1073/pnas.0905010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta S., Cano M., Ebrahimi K., Wang L., Handa J.T. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog. Retin. Eye Res. 2017;60:201–218. doi: 10.1016/j.preteyeres.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Lookeren Campagne M., LeCouter J., Yaspan B.L., Ye W. Mechanisms of age-related macular degeneration and therapeutic opportunities. J. Pathol. 2014;232:151–164. doi: 10.1002/path.4266. [DOI] [PubMed] [Google Scholar]
- 7.Gehrs K.M., Anderson D.H., Johnson L.V., Hageman G.S. Age-related macular degeneration—emerging pathogenetic and therapeutic concepts. Ann. Med. 2006;38:450–471. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bressler N.M. Age-related macular degeneration is the leading cause of blindness. JAMA. 2004;291:1900–1901. doi: 10.1001/jama.291.15.1900. [DOI] [PubMed] [Google Scholar]
- 9.Augustin A., Sahel J.A., Bandello F., Dardennes R., Maurel F., Negrini C., Hieke K., Berdeaux G. Anxiety and depression prevalence rates in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2007;48:1498–1503. doi: 10.1167/iovs.06-0761. [DOI] [PubMed] [Google Scholar]
- 10.Wong W.L., Su X., Li X., Cheung C.M., Klein R., Cheng C.Y., Wong T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob. Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell P., Liew G., Gopinath B., Wong T.Y. Age-related macular degeneration. Lancet. 2018;392:1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 12.Li W. Phagocyte dysfunction, tissue aging and degeneration. Ageing Res. Rev. 2013;12:1005–1012. doi: 10.1016/j.arr.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ach T., Tolstik E., Messinger J.D., Zarubina A.V., Heintzmann R., Curcio C.A. Lipofuscin redistribution and loss accompanied by cytoskeletal stress in retinal pigment epithelium of eyes with age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2015;56:3242–3252. doi: 10.1167/iovs.14-16274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Jiang C., Qin B., Liu G., Ji J., Sun X., Xu M., Ding S., Zhu M., Huang G. lncRNA ZNF503-AS1 promotes RPE differentiation by downregulating ZNF503 expression. Cell Death Dis. 2017;8:e3046. doi: 10.1038/cddis.2017.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulzbacher F., Kiss C., Kaider A., Eisenkoelbl S., Munk M., Roberts P., Sacu S., Schmidt-Erfurth U. Correlation of SD-OCT features and retinal sensitivity in neovascular age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2012;53:6448–6455. doi: 10.1167/iovs.11-9162. [DOI] [PubMed] [Google Scholar]
- 16.Li F., Wen X., Zhang H., Fan X. Novel insights into the role of long noncoding RNA in ocular diseases. Int. J. Mol. Sci. 2016;17:478. doi: 10.3390/ijms17040478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., Wu Z., Fu X., Han W. lncRNAs: insights into their function and mechanics in underlying disorders. Mutat. Res. Rev. Mutat. Res. 2014;762:1–21. doi: 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sen R., Ghosal S., Das S., Balti S., Chakrabarti J. Competing endogenous RNA: the key to posttranscriptional regulation. ScientificWorldJournal. 2014;2014:896206. doi: 10.1155/2014/896206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 23.Newman A.M., Gallo N.B., Hancox L.S., Miller N.J., Radeke C.M., Maloney M.A., Cooper J.B., Hageman G.S., Anderson D.H., Johnson L.V., Radeke M.J. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 2012;4:16. doi: 10.1186/gm315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curcio C.A., Presley J.B., Malek G., Medeiros N.E., Avery D.V., Kruth H.S. Esterified and unesterified cholesterol in drusen and basal deposits of eyes with age-related maculopathy. Exp. Eye Res. 2005;81:731–741. doi: 10.1016/j.exer.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Curcio C.A., Johnson M., Huang J.D., Rudolf M. Aging, age-related macular degeneration, and the response-to-retention of apolipoprotein B-containing lipoproteins. Prog. Retin. Eye Res. 2009;28:393–422. doi: 10.1016/j.preteyeres.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paraskevopoulou M.D., Hatzigeorgiou A.G. Analyzing miRNA-lncRNA interactions. Methods Mol. Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 27.Ballantyne M.D., McDonald R.A., Baker A.H. lncRNA/microRNA interactions in the vasculature. Clin. Pharmacol. Ther. 2016;99:494–501. doi: 10.1002/cpt.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J.Z., Shao C.C., Wang X.J., Zhao X., Chen J.Q., Ouyang Y.X., Feng J., Zhang F., Huang W.H., Ying Q. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175. doi: 10.1038/s41419-019-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muhammad N., Bhattacharya S., Steele R., Ray R.B. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016;7:58595–58605. doi: 10.18632/oncotarget.11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen M., Lechner J., Zhao J., Toth L., Hogg R., Silvestri G., Kissenpfennig A., Chakravarthy U., Xu H. STAT3 activation in circulating monocytes contributes to neovascular age-related macular degeneration. Curr. Mol. Med. 2016;16:412–423. doi: 10.2174/1566524016666160324130031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan B., Yao J., Liu J.Y., Li X.M., Wang X.Q., Li Y.J., Tao Z.F., Song Y.C., Chen Q., Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015;116:1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y., Dong L.F., Zhou R.M., Yao J., Song Y.C., Yang H., Jiang Q., Yan B. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: a clinical and in vitro study. J. Cell. Mol. Med. 2016;20:537–548. doi: 10.1111/jcmm.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J.J., Liu W.Q., Peng J.J., Ma Q.L., Peng J., Luo X.J. miR-21-5p/203a-3p promote ox-LDL-induced endothelial cell senescence through down-regulation of mitochondrial fission protein Drp1. Mech. Ageing Dev. 2017;164:8–19. doi: 10.1016/j.mad.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Han N., Xu H., Yu N., Wu Y., Yu L. miR-203a-3p inhibits retinal angiogenesis and alleviates proliferative diabetic retinopathy in oxygen-induced retinopathy (OIR) rat model via targeting VEGFA and HIF-1α. Clin. Exp. Pharmacol. Physiol. 2020;47:85–94. doi: 10.1111/1440-1681.13163. [DOI] [PubMed] [Google Scholar]
- 38.Chen L., Gao H., Liang J., Qiao J., Duan J., Shi H., Zhen T., Li H., Zhang F., Zhu Z., Han A. miR-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Am. J. Cancer Res. 2018;8:2387–2401. [PMC free article] [PubMed] [Google Scholar]
- 39.Fritsche L.G., Igl W., Bailey J.N., Grassmann F., Sengupta S., Bragg-Gresham J.L., Burdon K.P., Hebbring S.J., Wen C., Gorski M. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cascella R., Strafella C., Longo G., Ragazzo M., Manzo L., De Felici C., Errichiello V., Caputo V., Viola F., Eandi C.M. Uncovering genetic and non-genetic biomarkers specific for exudative age-related macular degeneration: significant association of twelve variants. Oncotarget. 2017;9:7812–7821. doi: 10.18632/oncotarget.23241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghanbari M., Erkeland S.J., Xu L., Colijn J.M., Franco O.H., Dehghan A., Klaver C.C.W., Meester-Smoor M.A. Genetic variants in microRNAs and their binding sites within gene 3'UTRs associate with susceptibility to age-related macular degeneration. Hum. Mutat. 2017;38:827–838. doi: 10.1002/humu.23226. [DOI] [PubMed] [Google Scholar]
- 42.Strafella C., Errichiello V., Caputo V., Aloe G., Ricci F., Cusumano A., Novelli G., Giardina E., Cascella R. The interplay between miRNA-related variants and age-related macular degeneration: evidence of association of MIR146A and MIR27A. Int. J. Mol. Sci. 2019;20:E1578. doi: 10.3390/ijms20071578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cascella R., Strafella C., Caputo V., Errichiello V., Zampatti S., Milano F., Potenza S., Mauriello S., Novelli G., Ricci F. Towards the application of precision medicine in age-related macular degeneration. Prog. Retin. Eye Res. 2018;63:132–146. doi: 10.1016/j.preteyeres.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 44.Eid R.A., Alkhateeb M.A., Eleawa S., Al-Hashem F.H., Al-Shraim M., El-Kott A.F., Zaki M.S.A., Dallak M.A., Aldera H. Cardioprotective effect of ghrelin against myocardial infarction-induced left ventricular injury via inhibition of SOCS3 and activation of JAK2/STAT3 signaling. Basic Res. Cardiol. 2018;113:13. doi: 10.1007/s00395-018-0671-4. [DOI] [PubMed] [Google Scholar]
- 45.Steyn P.J., Dzobo K., Smith R.I., Myburgh K.H. Interleukin-6 induces myogenic differentiation via JAK2-STAT3 signaling in mouse C2C12 myoblast cell line and primary human myoblasts. Int. J. Mol. Sci. 2019;20:E5273. doi: 10.3390/ijms20215273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wonganan O., He Y.J., Shen X.F., Wongkrajang K., Suksamrarn A., Zhang G.L., Wang F. 6-Hydroxy-3-O-methyl-kaempferol 6-O-glucopyranoside potentiates the anti-proliferative effect of interferon α/β by promoting activation of the JAK/STAT signaling by inhibiting SOCS3 in hepatocellular carcinoma cells. Toxicol. Appl. Pharmacol. 2017;336:31–39. doi: 10.1016/j.taap.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Liu K., Wu Z., Chu J., Yang L., Wang N. Promoter methylation and expression of SOCS3 affect the clinical outcome of pediatric acute lymphoblastic leukemia by JAK/STAT pathway. Biomed. Pharmacother. 2019;115:108913. doi: 10.1016/j.biopha.2019.108913. [DOI] [PubMed] [Google Scholar]
- 48.Brawley C., Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- 49.Xu C.H., Liu Y., Xiao L.M., Chen L.K., Zheng S.Y., Zeng E.M., Li D.H., Li Y.P. Silencing microRNA-221/222 cluster suppresses glioblastoma angiogenesis by suppressor of cytokine signaling-3-dependent JAK/STAT pathway. J. Cell. Physiol. 2019;234:22272–22284. doi: 10.1002/jcp.28794. [DOI] [PubMed] [Google Scholar]
- 50.Croker B.A., Kiu H., Nicholson S.E. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 2008;19:414–422. doi: 10.1016/j.semcdb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheng X.R., Brawley C.M., Matunis E.L. Dedifferentiating spermatogonia outcompete somatic stem cells for niche occupancy in the Drosophila testis. Cell Stem Cell. 2009;5:191–203. doi: 10.1016/j.stem.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alexander W.S., Hilton D.J. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu. Rev. Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- 53.Chen M., Zhao J., Ali I.H.A., Marry S., Augustine J., Bhuckory M., Lynch A., Kissenpfennig A., Xu H. Cytokine signaling protein 3 deficiency in myeloid cells promotes retinal degeneration and angiogenesis through arginase-1 up-regulation in experimental autoimmune uveoretinitis. Am. J. Pathol. 2018;188:1007–1020. doi: 10.1016/j.ajpath.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 54.Rawlings J.S., Rosler K.M., Harrison D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 55.Jiang C., Qin B., Liu G., Sun X., Shi H., Ding S., Liu Y., Zhu M., Chen X., Zhao C. MicroRNA-184 promotes differentiation of the retinal pigment epithelium by targeting the AKT2/mTOR signaling pathway. Oncotarget. 2016;7:52340–52353. doi: 10.18632/oncotarget.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.