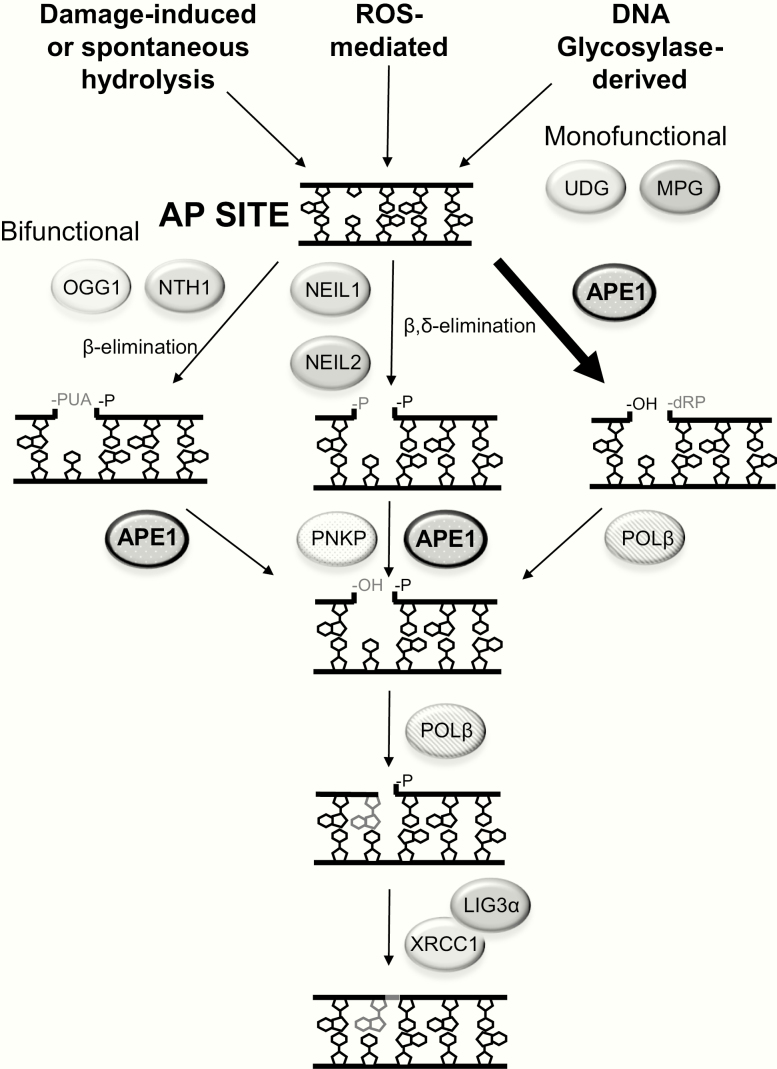
Figure 2.
AP site formation and repair. The three primary mechanisms for the generation of genomic AP sites are shown at the top, including BER-initiated via the activity of a monofunctional or bifunctional DNA glycosylase. Following AP site formation, the predominant repair process involves APE1 incision, creating a strand break with a 3′-hydroxyl (OH) and 5′-dRP (right). At some low frequency, bifunctional DNA glycosylases can cleave at the AP site via β-elimination (left) or β,δ-elimination (centre) to generate a strand break with a 3′-α,β-unsaturated aldehyde or 3′-phosphate (P), respectively and a 5′-P. The single-strand breaks are then processed by the depicted enzyme(s) to create 3′-OH and 5′-P single-nucleotide gap intermediates. Shown is single-nucleotide gap-filling, executed by DNA POLβ, and nick ligation by an XRCC1/LIG3α complex.