Abstract
Defects in DNA repair have been linked to the accumulation of somatic mutations in tumours. These mutations can promote oncogenesis; however, recent developments have indicated that they may also lead to a targeted immune response against the tumour. This response is initiated by the development of new antigenic epitopes (neoepitopes) arising from mutations in protein-coding genes that are processed and then presented on the surface of tumour cells. These neoepitopes are unique to the tumour, thus enabling lymphocytes to launch an immune response against the cancer cells. Immunotherapies, such as checkpoint inhibitors (CPIs) and tumour-derived vaccines, have been shown to enhance the immunogenic response to cancers and have led to complete remission in some cancer patients. There are tumours that are not responsive to immunotherapy or conventional tumour therapeutics; therefore, there is a push for new treatments to combat these unresponsive cancers. Recently, combinatorial treatments have been developed to further utilise the immune system in the fight against cancer. These treatments have the potential to exploit the defects in DNA repair by inducing more DNA damage and mutations. This can potentially lead to the expression of high levels of neoepitopes on the surface of tumour cells that will stimulate an immunological response. Overall, exploiting DNA repair defects in tumours may provide an edge in this long fight against cancer.
Introduction
Almost 50 years ago, Lawrence Loeb proposed that a mutator phenotype promotes oncogenesis and its progression (1,2). Current data support this hypothesis, and now it appears that this mutator phenotype can be exploited to combat cancer. Recently, studies have determined that neoepitopes derived from somatic mutations in tumours can be produced and may be presented on the surface of tumour cells. These neoepitopes may then be recognised by tumour infiltrating lymphocytes, enabling them to kill tumours. However, T cells eventually lose their capacity to invade and kill tumour cells. In order to elicit a potent immune response against tumour cells, immunotherapies have recently been developed to help boost the immune system.
Here, we review the link between mutation burden and immunotherapy in addition to how the DNA repair landscape impacts the formation of neoepitopes. We then discuss the potential role of DNA damaging chemotherapeutic agents in the induction of neoepitopes, especially in DNA repair defective cells.
Immunotherapy and Mutation Burden
The ability of the immune system to suppress tumour growth is dependent on two factors: the immunogenicity of the tumour and the immune response. The first evidence that tumour cells may be immunogenic and that an immune response can be mounted against tumours is found in studies that show that mice can be immunised with syngeneic tumours (3–5). Further research indicates that immunisation by one tumour does not elicit an immune response against other tumours of the same type, suggesting that there are tumour-specific immunostimulatory elements (4,6). Accordingly, the abundance of tumour infiltrating lymphocytes (TILs), such as T cells, has been associated with good clinical outcome in patients with primary solid tumours (7–9). However, there can be continued tumour progression despite high infiltration of T cells (8). This can be due to T cells becoming functionally unresponsive (anergy) through T-cell tolerance (reviewed here (10)). Another mechanism that would allow continued tumour progression involves T cells eventually losing their functional capacities to secrete effector proteins, proliferate, and lyse cells upon antigen exposure. This ‘T-cell exhaustion’ (in-depth reviews on T-cell exhaustion can be found here: (11–13)) can further lead to the upregulation of inhibitory receptors, such as program death 1 (PD-1), cytotoxic T lymphocyte antigen 4 (CTLA-4), and lymphocyte activation gene 3 (LAG-3) (14–16). Collectively, these dysfunctions in T cells contribute to the inability of the body to mount an optimal immune response against tumours, even if the tumour is immunogenic.
To jumpstart the immune system, immunotherapies have been developed to enhance immunogenic responses toward cancers. Currently, the different types of immunotherapies include checkpoint inhibitors (CPIs) (17–19), adoptive cell transfer (20), cytokines (21), tumour-derived vaccines (22), and bacteria-derived vaccines (23) (the immunotherapy historical context can be appreciated here: (24)). In this section, we will focus on the development and modes of action of CPIs and the association between CPI efficacy and the total number of mutations in a tumour genome, which is also termed as tumour mutation burden (TMB).
The development of CPIs was based on work that determined the functional mechanism of how inhibitory T-cell receptor proteins halt T-cell activation and their ability to kill tumour cells. Early work in this field characterised two inhibitory T-cell receptors, CTLA-4 and PD-1 (25–27). In 1996 it was shown for the first time that the use of antibodies against the inhibitory receptor CTLA-4 could enhance anti-tumour immunity in vivo (28). The studies of CTLA-4 and PD-1 eventually led to two cancer immunologists, James Allison and Tasuku Honjo, being honoured in 2018 with the Nobel Prize in Medicine. Their work, in addition to that of many other researchers, resulted in the use of monoclonal antibodies as CPIs to target specific inhibitory receptors or their cognate partners. This permits reinvigoration of T cells from their exhaustive state and thus stimulates an immune response that leads to infiltration of lymphocytes into the tumour and the eventual death of tumour cells (Figure 1). Currently, there are several CPIs, including anti-CTLA-4, anti-PD-1, and anti-PD-L1 (the ligand to PD-1), that have been approved by the Food and Drug Administration in the last two decades. In the coming years, many more CPIs may be approved as there are currently numerous studies analysing other checkpoint proteins, such as LAG3 (29), T-cell membrane protein 3 (TIM3) (30), or even certain metabolic enzymes, such as indoleamine 2,3-dioxygenase (IDO) (31).
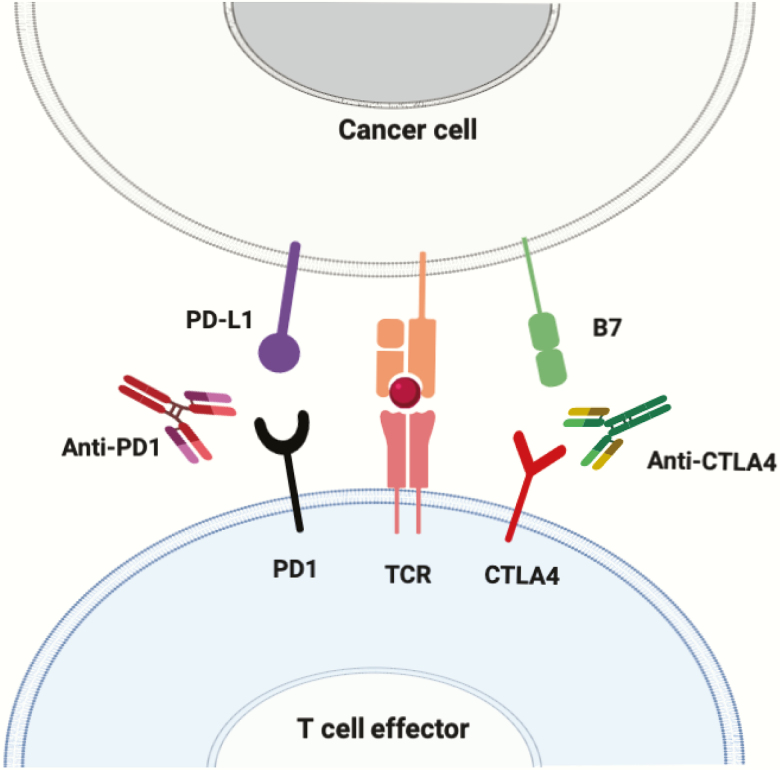
Figure 1.
Immune checkpoint evasion. Neoepitopes expressed in tumour cells are presented on the Major Histocompatibility Complex (MHC). This eventually leads to the interaction of neoepitopes with their cognate T-cell receptor (TCR) located on effector T cells. If checkpoint ligands are not expressed on the tumour, the T cells will secrete effector proteins and proliferate, initiating an immune response that is directed toward the tumour cells. However, if the tumour cells express checkpoint ligands (e.g. PD-L1 and B7), upon MHC-neoepitope/TCR interaction, the checkpoint ligands will further interact with their cognate partners (e.g. PD1 and CTLA4, respectively) and inhibit the effector function of the T cells. Monoclonal antibodies to the checkpoint ligands or receptors remove this checkpoint and promotes T-cell effector functions. Figure available in colour online.
Current CPIs are effective for some tumours and show no response in other tumours. To better target cancer cells, biomarkers are being analysed to determine which ones strongly correlate with CPI responses. One of the main biomarkers that is associated with CPI efficacy is a high, non-synonymous TMB (32–36). Additionally, CPI efficacy is correlated with mutations in DNA repair genes, including the mismatch repair (MMR) gene mut S homolog 2 (MSH2) and the non-homologous end-joining (NHEJ) gene DNA dependent protein kinase catalytic subunit (PRKDC), likely due to the higher levels of base substitutions and insertions and deletions (indels) in tumour cells defective in DNA repair (33,37). High TMB is also correlated with the molecular smoking signature in non-small cell lung cancers (NSCLC), as a result of massive levels of DNA damage that are not all correctly repaired, leading to inflammation and increased mutation burden (33). Multiple clinical trials analysing melanoma, NSCLC, urothelial cancer, and squamous cell carcinomas have established a mutation threshold indicative of beneficial results with CPI therapy. For example, in an NSCLC clinical trial, patients with >10 mutations per megabase show a progression-free survival of 7.1 versus 3.2 months in patients with tumour mutation levels <10 mutations per megabase (38) (refer to (36) for a list of clinical trials that set TMB threshold for CPI benefit). Interestingly, tumours that have the best responses to CPIs are environmentally associated cancers, such as melanoma and lung cancer. This may be because UV light and cigarette smoke are known to induce DNA damage, which, if repaired incorrectly, can result in mutations (39). The association between DNA repair and environmental carcinogens has been reaffirmed with the finding that cancer mutation signatures associated with environmental agents are linked to the dysfunction of various repair pathways, including MMR, transcription-coupled nucleotide excision repair (NER), and direct reversal repair pathways (40).
The Mutator Phenotype and Cancer
In 1974, it was proposed that DNA polymerase infidelity during DNA replication promoted a mutator phenotype that was responsible for tumourigenesis and progression of cancers (1). This mutator phenotype hypothesis was later amended to include defects in DNA repair, where it was posited that DNA repair defects could lead to DNA damage exceeding the DNA repair capacity of the cell and any residual unrepaired lesions would result in mutations after replication (2). As tumour sequencing data accumulated in The Cancer Genome Atlas, it became apparent that tumours had tens to hundreds of thousands of mutations (41,42), suggesting that the mutator phenotype hypothesis was correct. In this section, we will focus on the mutator phenotypes that result from mutations in DNA polymerase genes and DNA repair genes in tumours (Figure 2).
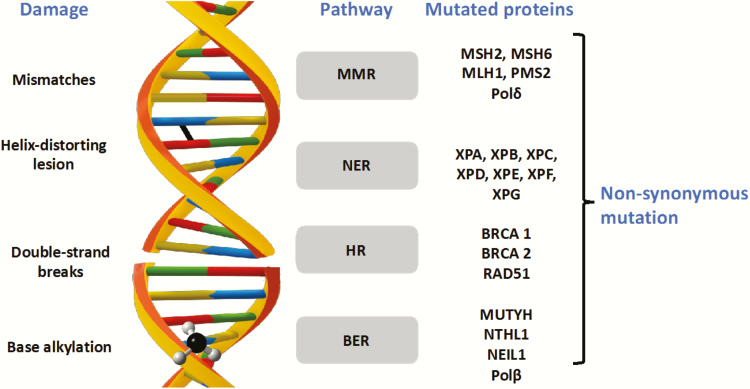
Figure 2.
DNA repair pathways and the proteins mutated in cancer. Lesion-specific DNA repair mechanisms and the proteins that are frequently mutated in cancer are shown. Figure available in colour online.
DNA polymerases
During DNA replication, it has been estimated that an error occurs every 109–1010 nucleotides. This low mutation frequency is dependent on MMR as well as the proofreading exonuclease domains of DNA polymerases (Pol) δ and ε (43,44). Studies in mice demonstrate that defective 3′ to 5′ exonuclease activity of either Polδ or Polε results in a strong spontaneous mutator phenotype and increased tumour incidence (45,46). Remarkably, germline mutations in the exonucleolytic proofreading domains of Polδ and Polε predispose patients to colorectal adenomas and carcinomas (47–50). Somatic mutations in POLD1 and POLE catalytic subunits have also been frequently reported in colorectal and endometrial tumours and are occasionally found in ovarian, breast, brain, prostrate, pancreas, and lung tumours (51–53). Interestingly, the POLE exonuclease domain is mutated in ~12.3% of microsatellite-stable sporadic colorectal cancers (CRCs) with over 50 mutations per megabase (54,55). Recently, it has been found that the Polε-P286R variant amplifies its polymerase activity at the expense of its exonuclease activity, resulting in an ultramutator phenotype (56). Additionally, mutations in Polη, a translesion synthesis polymerase, can result in a high mutation frequency due to impaired bypass of thymine dimers, the most common lesion generated in DNA by ultraviolet (UV) radiation, leading to skin cancer (57).
Base excision repair
Base excision repair (BER) recognises and repairs non-helix distorting lesions, such as oxidised bases and single-strand breaks. During BER, lesion-specific monofunctional DNA glycosylases (e.g. MUTYH) recognise and excise damaged bases from the phosphate backbone, resulting in an apurinic/apyrimidinic (AP) site. The apurinic/apyrimidinic endonuclease 1 (APE1) then nicks the DNA backbone 5′ of the AP site. Bifunctional glycosylases (e.g. nth like DNA glycosylase 1 (NTHL1)) can additionally cleave the backbone, leaving blocked ends that must be enzymatically remodelled (58,59). Afterwards, DNA polymerase β (Polβ) incorporates a single nucleotide into the gap and removes the 5′-deoxyribose phosphate. Finally, ligases I or IIIα seal the nick (60).
Germline biallelic mutations in MUTYH, impair the removal of adenine opposite 8-oxo-guanine, one of the most abundant oxidative lesions. This leads to an increased frequency of G:C➔T:A transversions and MUTYH-associated polyposis (MAP). MAP patients have high frequencies of somatic mutations in the APC and KRAS genes (61–64) and a 28-fold lifetime risk of CRC (65). Homozygous germline mutations in NTHL1 lead to adenomatous polyposis and CRC (66). Interestingly, somatic mutations in CRC driver genes, KRAS, APC, TP53 and PIK3CA, along with C:G➔T:A transitions, were identified in carcinomas carrying nonsense mutations in NTHL1 encoding p.Gln90*, primarily due to the inability of the mutant protein to remove oxidised pyrimidines from DNA. NTHL1 mutations are also associated with multiple malignancies, including bladder cancer, basal cell carcinoma, and endometrial breast cancers, although the presence of NTHL1 mutations are not associated with a mutator phenotype (66).
Our lab has shown that germline BER variants POLB P242R (67), NTHL1 D239Y (68), and NEIL1 G83D (69), are capable of inducing genomic instability in cells. In summary, mutations in BER genes are linked to increased levels of mutagenesis due to their inability to repair DNA damage. Therefore, this suggests that tumours harbouring mutations in BER genes may produce increased levels of neoepitopes.
Nucleotide excision repair
Over 30 NER proteins work together in a series of steps to remove helix-distorting DNA lesions that arise from exogenous sources, such as tobacco smoke and UV radiation (70). Briefly, proteins such as xeroderma pigmentosum complementation group E (XPE), DNA damage-binding protein 1 (DDB1), XPC, RAD23 homologue B (RAD23B), and centrin-2 recognise the damage (71). Next, DNA unwinding and dual incisions 3′ and 5′ of the DNA adduct release an oligonucleotide of ~25–30 bases. This is followed by gap-filling and finally, DNA ligation (72). Defects in the NER pathway lead to an autosomal recessive disorder called xeroderma pigmentosum (XP). Tumours from XP patients show an increase in mutation frequencies in the Ras family of proto-oncogenes and the tumour suppressor gene p53 compared to the normal population (73). Thus, it is speculated that tumours associated with mutations in NER genes may induce increased levels of neoepitopes.
Double-strand break repair
Double-strand breaks (DSBs) are the most toxic lesions for cells and can arise from collapsed replication forks or ionising radiation. The initial recognition of DSBs is by the meiotic recombination 11 homolog 1 (MRE11) complex (MRE11, Nijmegen Breakage Syndrome 1(NBS1), and RAD50 DSB Repair Protein (RAD50)), which catalyses the activation of ataxia-telangiectasia mutated protein (ATM) in conjunction with tat-interactive protein 60 kDa (TIP60) and p53-binding protein (53BP1) (74). Afterwards, resection by the MRE11 complex and C-terminal-binding protein interacting protein (CtIP) dictate the pathway choice for repair of the DSB. These pathways include homologous recombination (HR), non-homologous end-joining (NHEJ), and alternative NHEJ (alt-NHEJ) (75). In HR, homologous DNA sequences are used to repair DSBs, while the error-prone NHEJ pathway ligates the DSB ends together directly, without extensive homology. Therefore, NHEJ is associated with small insertions and deletions of several base pairs at the break site (76).
HR-deficient cells, such as those found in carriers of BRCA and RAD51 mutations, showed an increased cancer risk across multiple clinical studies (77). Women with inherited germline BRCA1 or BRCA2 mutations have a cumulative lifetime risk of ~72 and 69%, respectively, for developing breast cancers (78). In vivo mouse studies demonstrate that somatic loss of BRCA2 causes a 2.3-fold increase in mutation burden, which is equivalent to an extra 100 mutations per cell (79). Intriguingly, mutations in BRCA1/2 are associated with an increased mutation load and higher levels of neoepitopes compared to HR proficient ovarian cancer tumours (80). In the absence of HR, low fidelity DSB repair pathways, such as NHEJ and alternative NHEJ (alt-NHEJ), lead to the accumulation of point mutations and random indels resulting in high mutational burden and neoepitope expression (81,82).
Mismatch repair
MMR recognises and repairs mispaired bases and indels that arise from normal DNA processes, such as DNA replication, recombination, and repair. The two major MMR complexes that recognise these lesions are MutSα, a heterodimer of Mut S Homolog 2 (MSH2) and Mut S Homolog 6 (MSH6) that recognises mispairs and short indel loops (1–2 nucleotides) (83), and MutSβ, a heterodimer of MSH2 and Mut S Homolog 3 (MSH3) that recognises long indel loops (1–20 nucleotides) (84–86). After recognition of the lesion, the MutSα or MutSβ complex uses its ATPase activity to convert itself into a sliding clamp that then recruits MutLα, which is comprised of MLH1 and PMS2, to nick the DNA. Exonuclease 1 then resects the DNA, producing a large DNA gap that is then filled in by Polδ (43).
Defective MMR increases mutation rates up to 1000-fold, which results in microsatellite instability (MSI) and is associated with cancer development (87). Patients with germline mutations in MMR genes (e.g., MSH2, MSH6, MLH1, and Post-meiotic segregation 2 (PMS2)) develop Lynch syndrome and have a greater risk of developing CRC and endometrial cancer (88). Additionally, somatic mutations in MLH1 and MSH2 were identified in MSI-positive tumours (89), and biallelic germline mutations in MSH3 are linked to colorectal adenomatous polyposis (90). MLH1 inactivation is also associated with a high mutational load and increased levels of neoepitopes (91).
In summary, tumours with defects in various DNA repair pathways have increased levels of mutations. Although these mutations play a role in driving carcinogenesis, they may also increase the levels of tumour neoepitopes. Therefore, tumours with mutations in DNA repair genes may respond to CPIs.
The Mutator Phenotype and Neoepitopes
Somatic mutations contribute to the development of cancers; however, these mutations can also be leveraged to inhibit cancer growth. Aberrant peptide sequences encoded by somatic mutations in the tumour can lead to the presentation of neoepitopes on the cell surface by the Major Histocompatibility Complex (MHC), where they are subjected to immunosurveillance. The collection of cell-surface antigenic peptides, known as the immunopeptidome, is generated by a combination of intracellular processes known as the antigen presentation pathway. This pathway dictates the presentation of tumour-specific neoepitopes that can differentiate between a host’s normal tissue and neoplastic tissue.
The antigen presentation pathway comprises several key steps: proteasomal digestion of intracellular proteins, transportation into the endoplasmic reticulum (ER), conjugation with MHC class I (MHC I) molecules, and vesicular trafficking to the cell surface. A protein destined for degradation is tagged with a poly-ubiquitin chain, causing it to be targeted by the proteasome complex where it is cleaved into smaller peptide sequences ranging from 8 to 30 amino acids (92). Many of these short peptide sequences will be degraded by aminopeptidases present in the cytosol (>99%); however, a small fraction will be channelled into the ER space by the transporter associated with antigen processing (TAP) complex. Peptides sequestered into the ER space may undergo further modification from aminopeptidases before potentially binding to MHC I molecules (92). Once bound to MHC I molecules, peptides are enveloped and trafficked by the Golgi Apparatus to be presented on the cell surface (Figure 3). It is at the interface between the cell surface and the extracellular space where T cells can interact and form a ternary complex with peptide-MHC conjugates. As neoepitopes are derived from tumour-specific mutations that deviate from the host’s immunopeptidome, CD8+ T cells initiate clearance of tumourigenic cells based on neoepitope detection.
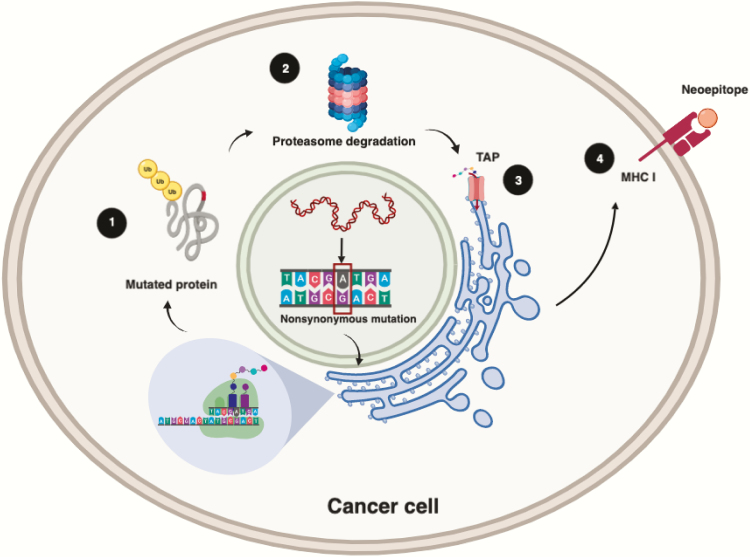
Figure 3.
Neoepitope presentation. The presentation of neoepitopes begins with the ubiquitylation and proteasome degradation of mutated proteins into short polypeptides (1 and 2). These smaller peptides enter the ER through the TAP complex (3). Once in the ER, the peptides bind to MHC class I. Together, the peptide–MHC class I complexes are transported to the cell membrane by the Golgi complex (4) where the peptide can be recognised by T cells. Figure available in colour online. Figure created with BioRender.com.
The molecular landscape of mutations in tumours (mutanome) has been made possible with the advent of genomic sequencing. Consequently, computational tools are being developed to identify viable mutanome-derived neoepitopes for immunotherapy. This has sparked an area of intensive research for neoepitope prediction that involves modelling the binding affinity of peptide sequences to MHC alleles. Modelling this biophysical interaction is of great interest as the peptide-MHC binding step has been used as the most selective aspect in the antigenic selection process (93). However, recent studies indicate that the binding affinity of the peptide to MHC may not be the most efficient way to select the right antigen (94). As the MHC loci are highly polymorphic, there is a panoply of MHC molecules that can be generated. This proteomic diversity increases the potential combinations of antigenic peptides and MHC conjugates that can be bound together. As a result, a multitude of prediction algorithms have been developed solely for modelling peptide-MHC binding. Though nascent approaches require a wealth of peptide-MHC allele binding data and are only able to model a few allelic variants, advancements in MHC-binding prediction algorithms have improved affinity predictions with smaller training datasets and broadened prediction to MHC alleles without explicit allelic binding data (95). Mass spectrometry is being performed on the MHC ligandome (immune-peptidome) in combination with genomic sequencing. These approaches have already been successfully utilised in several studies (96,97). Continued development of bioinformatic and biophysical approaches will further inform each discipline, augment the understanding of the immunopeptidome, and enhance immunotherapeutic treatment regimens (97).
Combining DNA Repair Defects and Drugs to Induce Additional Neoepitopes
DNA repair is crucial in preventing tumourigenesis and maintaining genomic stability. Cancer cells frequently have reduced DNA repair as a result of mutations in DNA repair genes and are often more susceptible to DNA damage (98). Chemotherapies (e.g. platinum-based drugs, alkylating agents, and DNA intercalators) and radiotherapies can damage DNA, which, if left unrepaired in tumours, can lead to cell death and/or an increase in TMB. Based on studies discussed in previous sections, it is evident that DNA repair deficiency is highly associated with increased neoepitopes in tumours. Therefore, treatment of DNA repair-deficient tumour cells with DNA damaging agents may induce an increase in the levels of neoepitopes that, when combined with immunotherapy, can facilitate a potent recruitment and response by the immune system to the tumour (99). Recent evidence suggests that tumours with high levels of neoepitopes present an improved response to CPIs, while the loss of neoepitopes is associated with CPI resistance (100,101). Ongoing clinical trials are studying these combinatorial treatments to determine their efficacy (Table 1). Thus, combinatorial therapies, in which DNA damaging chemotherapeutics are used together with immunotherapeutic agents, can synergise with the therapeutic potential of anticancer drugs (102,103). The CPI’s response has been correlated with TMB and the levels of neoepitopes. However, mutational load alone is not enough to drive a CPI’s response, and a critical challenge is to identify key mutations that may result in specific types of neoepitopes that have the capacity to drive immune response.
Table 1.
Combining DNA damaging chemotherapeutics with immunotherapeutics
| DNA damaging chemotherapy | DNA lesions | DNA repair | In combination with | Reference |
|---|---|---|---|---|
| Platinum-based (Cisplatin) | Monofunctional DNA adducts, DNA-protein crosslinks, intrastrand and interstrand crosslinks | NER (major), MMR, HR, BER, NHEJ | PARPi (Olaparib) | (104) |
| Cisplatin | Anti-PD-1 (pembrolizumab) | (105) | ||
| Alkylating agents (Temozolamide) | N7 and O6 methyl guanine adducts, interstrand crosslinks | MGMT, NER | IFN alpha 2B | (106,107) |
| Microwave | Anti-CTLA-4 (tremelimumab) | (108) | ||
| Ionising radiation | DSB (major), ROS, SSB | NHEJ (major) | PD-L1 inhibitor (durvalumab) | (109,110) |
Tumours with mutations in DNA repair genes show a positive response to immunotherapy treatments (35,99). For instance, using PARP inhibition and anti-PD1 or PD-L1 to promote TILs in patients’ samples may be a promising combinatorial therapy that takes advantage of defective DNA repair in cells harbouring BRCA mutations (111). Genomic instability induced by a defective DNA repair response has significant potential to result in increased tumour mutational burden and increased levels of neoepitopes that could further contribute to CPI sensitivity.
Conclusions/Future Directions
Over the last few decades, there have been many discoveries that have enabled immunotherapy to be used in cancer treatment. Mutations arising from aberrant DNA polymerases and DNA repair genes lead to the production of neoepitopes that are unique to the tumour. Immunotherapy and combinatorial therapies can be used to stimulate the immune system to kill cancer cells. These treatments have proven effective for some cancers; however, even with all these advancements, some cancer types remain unresponsive.
Biomarkers, such as the TMB, have become important in identifying responders to CPIs. Currently, there are other biomarkers being discovered that correlate with CPI efficacy in patients. One of these biomarkers is the quality of neoepitopes (32–36). It has been observed that some mutations, such as those found in a viral open reading frame in the cancer’s genome, result in antigens that are of a higher quality than other antigens (112). The neoepitopes that arise are more easily recognised as ‘non-self’ by the immune system and thus a stronger immune response is launched. Another study determined that the quantity of neoantigens alone was not enough to predict long-term survivors of pancreatic cancer; however, a model that attributed greater immunogenicity to neoepitopes that were homologous to infectious disease-derived peptides and had differential presentation was able to successfully identify long-term survivors in two independent datasets (113,114).
Immunotherapy treatments would also benefit from creating a higher quantity of neoepitopes by further impeding various DNA repair pathways. This review has already discussed the use of DNA damaging agents to promote higher neoepitope quantity. Another way to achieve this is through epigenetic silencers of DNA repair genes, as it was found that silencing repair genes through epigenetic processes was associated with a high mutational burden (115).
While countless cancer patients have already benefited from the use of immunotherapy in cancer treatments, many more have cancers that remain unresponsive to treatment. Further advancements hold the potential to help these patients by continuing to use one of the hallmarks of cancer against itself.
Funding
This work was supported by 1R21 CA216595 from that National Cancer Institute to JBS.
Conflict of interest statement
None declared.
References
- 1. Loeb L. A., Springgate C. F. and Battula N (1974) Errors in DNA replication as a basis of malignant changes. Cancer Res., 34, 2311–2321. [PubMed] [Google Scholar]
- 2. Loeb L. A. (2001) A mutator phenotype in cancer. Cancer Res., 61, 3230–3239. [PubMed] [Google Scholar]
- 3. Gross L. (1943) Intradermal Immunization of C3H Mice against a Sarcoma That Originated in an Animal of the Same Line. Cancer Res., 3, 326–333. [Google Scholar]
- 4. Prehn R.T. and Main J. M (1957) Immunity to methylcholanthrene-induced sarcomas. J. Natl. Cancer Inst., 18, 769–778. [PubMed] [Google Scholar]
- 5. Klein G., Sjogren H. O., Klein E. and Hellstrom K. E (1960) Demonstration of resistance against methylcholanthrene-induced sarcomas in the primary autochthonous host. Cancer Res., 20, 1561–1572. [PubMed] [Google Scholar]
- 6. Basombrío M. A. (1970) Search for common antigenicities among twenty-five sarcomas induced by methylcholanthrene. Cancer Res., 30, 2458–2462. [PubMed] [Google Scholar]
- 7. Zhang L., Conejo-Garcia J. R., Katsaros D., et al. (2003) Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med., 348, 203–213. [DOI] [PubMed] [Google Scholar]
- 8. Azimi F., Scolyer R. A., Rumcheva P., Moncrieff M., Murali R., McCarthy S. W., Saw R. P. and Thompson J. F (2012) Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J. Clin. Oncol., 30, 2678–2683. [DOI] [PubMed] [Google Scholar]
- 9. Galon J., Costes A., Sanchez-Cabo F., et al. (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science, 313, 1960–1964. [DOI] [PubMed] [Google Scholar]
- 10. Boon T., Coulie P. G., Van den Eynde B. J. and van der Bruggen P (2006) Human T cell responses against melanoma. Annu. Rev. Immunol., 24, 175–208. [DOI] [PubMed] [Google Scholar]
- 11. Abe B. T. and Macian F (2013) Uncovering the mechanisms that regulate tumor-induced T-cell anergy. Oncoimmunology, 2, e22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schietinger A. and Greenberg P. D (2014) Tolerance and exhaustion: defining mechanisms of T cell dysfunction. Trends Immunol., 35, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang Y., Li Y. and Zhu B (2015) T-cell exhaustion in the tumor microenvironment. Cell Death Dis., 6, e1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Day C. L., Kaufmann D. E., Kiepiela P., et al. (2006) PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature, 443, 350–354. [DOI] [PubMed] [Google Scholar]
- 15. Woo S. R., Turnis M. E., Goldberg M. V., et al. (2012) Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res., 72, 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosenberg S. A., Sherry R. M., Morton K. E., et al. (2005) Tumor progression can occur despite the induction of very high levels of self/tumor antigen-specific CD8+ T cells in patients with melanoma. J. Immunol., 175, 6169–6176. [DOI] [PubMed] [Google Scholar]
- 17. Havel J. J., Chowell D. and Chan T. A (2019) The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer, 19, 133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei S. C., Duffy C. R. and Allison J. P (2018) Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov., 8, 1069–1086. [DOI] [PubMed] [Google Scholar]
- 19. Lee A., Sun S., Sandler A. and Hoang T (2018) Recent progress in therapeutic antibodies for cancer immunotherapy. Curr. Opin. Chem. Biol., 44, 56–65. [DOI] [PubMed] [Google Scholar]
- 20. Rosenberg S. A. and Restifo N.P. (2015) Adoptive cell transfer as personalized immunotherapy for human cancer. Science, 348, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Conlon K. C., Miljkovic M. D. and Waldmann T. A (2019) Cytokines in the treatment of cancer. J. Interferon Cytokine Res., 39, 6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu Z., Ott P. A. and Wu C. J (2018) Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat. Rev. Immunol., 18, 168–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaimala S., Al-Sbiei A., Cabral-Marques O., Fernandez-Cabezudo M. J. and Al-Ramadi B. K (2018) Attenuated bacteria as immunotherapeutic tools for cancer treatment. Front. Oncol., 8, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oiseth S. and Aziz M.S (2017) Cancer immunotherapy: a brief review of the history, possibilities, and challenges ahead. J. Cancer Metastasis Treat., 3, 250–261. [Google Scholar]
- 25. Waterhouse P., Penninger J. M., Timms E., Wakeham A., Shahinian A., Lee K. P., Thompson C. B., Griesser H. and Mak T. W (1995) Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science, 270, 985–988. [DOI] [PubMed] [Google Scholar]
- 26. Tivol E. A., Borriello F., Schweitzer A. N., Lynch W. P., Bluestone J. A. and Sharpe A. H (1995) Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity, 3, 541–547. [DOI] [PubMed] [Google Scholar]
- 27. Ishida Y., Agata Y., Shibahara K. and Honjo T (1992) Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J., 11, 3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leach D. R., Krummel M. F. and Allison J. P (1996) Enhancement of antitumor immunity by CTLA-4 blockade. Science, 271, 1734–1736. [DOI] [PubMed] [Google Scholar]
- 29. Andrews L. P., Marciscano A. E., Drake C. G. and Vignali D. A (2017) LAG3 (CD223) as a cancer immunotherapy target. Immunol. Rev., 276, 80–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson A. C. (2014) Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol. Res., 2, 393–398. [DOI] [PubMed] [Google Scholar]
- 31. Yentz S. and Smith D (2018) Indoleamine 2,3-Dioxygenase (IDO) inhibition as a strategy to augment cancer immunotherapy. BioDrugs, 32, 311–317. [DOI] [PubMed] [Google Scholar]
- 32. Snyder A., Makarov V., Merghoub T., et al. (2014) Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med., 371, 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rizvi N. A., Hellmann M. D., Snyder A., et al. (2015) Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science, 348, 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carbone D. P., Reck M., Paz-Ares L., et al. ; CheckMate 026 Investigators (2017) First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med., 376, 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hugo W., Zaretsky J. M., Sun L., et al. (2016) Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell, 165, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chan T. A., Yarchoan M., Jaffee E., Swanton C., Quezada S. A., Stenzinger A. and Peters S (2019) Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann. Oncol., 30, 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mandal R., Samstein R. M., Lee K. W., et al. (2019) Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science, 364, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hellmann M. D., Ciuleanu T. E., Pluzanski A., et al. (2018) Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med., 378, 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schumacher T. N. and Schreiber R. D (2015) Neoantigens in cancer immunotherapy. Science, 348, 69–74. [DOI] [PubMed] [Google Scholar]
- 40. Kucab J. E., Zou X., Morganella S., et al. (2019) A compendium of mutational signatures of environmental agents. Cell, 177, 821–836.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Greenman C., Stephens P., Smith R., et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature, 446, 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Loeb L. A. (2011) Human cancers express mutator phenotypes: origin, consequences and targeting. Nat. Rev. Cancer, 11, 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kunkel T. A. and Erie D. A (2005) DNA mismatch repair. Annu. Rev. Biochem., 74, 681–710. [DOI] [PubMed] [Google Scholar]
- 44. Bielas J. H. and Loeb L. A (2005) Mutator phenotype in cancer: timing and perspectives. Environ. Mol. Mutagen., 45, 206–213. [DOI] [PubMed] [Google Scholar]
- 45. Albertson T. M., Ogawa M., Bugni J. M., et al. (2009) DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl. Acad. Sci. U. S. A., 106, 17101–17104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goldsby R. E., Lawrence N. A., Hays L. E., Olmsted E. A., Chen X., Singh M. and Preston B. D (2001) Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat. Med., 7, 638–639. [DOI] [PubMed] [Google Scholar]
- 47. Palles C., Cazier J. B., Howarth K. M., et al. ; CORGI Consortium; WGS500 Consortium (2013) Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet., 45, 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith C. G., Naven M., Harris R., et al. (2013) Exome resequencing identifies potential tumor-suppressor genes that predispose to colorectal cancer. Hum. Mutat., 34, 1026–1034. [DOI] [PubMed] [Google Scholar]
- 49. Dunlop M. G., Dobbins S. E., Farrington S. M., et al. ; Colorectal Tumour Gene Identification (CORGI) Consortium; Swedish Low-Risk Colorectal Cancer Study Group; COIN Collaborative Group (2012) Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat. Genet., 44, 770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cancer Genome Atlas Network, T.C.G.A. (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cerami E., Gao J., Dogrusoz U., et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Forbes S. A., Beare D., Gunasekaran P., et al. (2015) COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res., 43, D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barbari S. R. and Shcherbakova P. V (2017) Replicative DNA polymerase defects in human cancers: consequences, mechanisms, and implications for therapy. DNA Repair (Amst)., 56, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guerra J., Pinto C., Pinto D., et al. (2017) POLE somatic mutations in advanced colorectal cancer. Cancer Med., 6, 2966–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stenzinger A., Pfarr N., Endris V., et al. (2014) Mutations in POLE and survival of colorectal cancer patients–link to disease stage and treatment. Cancer Med., 3, 1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xing X., Kane D. P., Bulock C. R., Moore E. A., Sharma S., Chabes A. and Shcherbakova P. V (2019) A recurrent cancer-associated substitution in DNA polymerase ε produces a hyperactive enzyme. Nat. Commun., 10, 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kunkel T. A., Pavlov Y. I. and Bebenek K (2003) Mini review Functions of human DNA polymerases, and suggested by their properties, including fidelity with undamaged DNA templates.DNA Repair, 2, 135–149. [DOI] [PubMed] [Google Scholar]
- 58. Krokan H. E. and Bjørås M (2013) Base excision repair. Cold Spring Harb. Perspect. Biol., 5, a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wallace S. S., Murphy D. L. and Sweasy J. B (2012) Base excision repair and cancer. Cancer Lett., 327, 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fleck O. and Nielsen O (2004) DNA repair. J. Cell Sci., 117, 515–517. [DOI] [PubMed] [Google Scholar]
- 61. Mork M. E. and Vilar E (2016) MUTYH-associated polyposis. Intestinal polyposis syndromes: diagnosis and management. Elsevier, 79, 25–32. [Google Scholar]
- 62. David S. S., O’Shea V. L. and Kundu S (2007) Base-excision repair of oxidative DNA damage. Nature, 447, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sampson J. R., Jones S., Dolwani S. and Cheadle J. P (2005) MutYH (MYH) and colorectal cancer. Biochem. Soc. Trans., 33, 679–683. [DOI] [PubMed] [Google Scholar]
- 64. Lipton L., Halford S. E., Johnson V., et al. (2003) Carcinogenesis in MYH-associated polyposis follows a distinct genetic pathway. Cancer Res., 63, 7595–7599. [PubMed] [Google Scholar]
- 65. Ferlay J., Steliarova-Foucher E., Lortet-Tieulent J., Rosso S., Coebergh J. W., Comber H., Forman D. and Bray F (2013) Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur. J. Cancer, 49, 1374–1403. [DOI] [PubMed] [Google Scholar]
- 66. Weren R. D., Ligtenberg M. J., Kets C. M., et al. (2015) A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat. Genet., 47, 668–671. [DOI] [PubMed] [Google Scholar]
- 67. Yamtich J., Nemec A. A., Keh A. and Sweasy J. B (2012) A germline polymorphism of DNA polymerase beta induces genomic instability and cellular transformation. PLoS Genet., 8, e1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Galick H. A., Kathe S., Liu M., Robey-Bond S., Kidane D., Wallace S. S. and Sweasy J. B (2013) Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proc. Natl. Acad. Sci. U. S. A., 110, 14314–14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Galick H. A., Marsden C. G., Kathe S., et al. (2017) The NEIL1 G83D germline DNA glycosylase variant induces genomic instability and cellular transformation. Oncotarget, 8, 85883–85895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shell S. M. and Chazin W. J (2012) XPF-ERCC1: on the bubble. Structure, 20, 566–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hanawalt P. C. and Spivak G (2008) Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol., 9, 958–970. [DOI] [PubMed] [Google Scholar]
- 72. Sancar A. and Tang M. S (1993) Nucleotide excision repair. Photochem. Photobiol., 57, 905–921. [DOI] [PubMed] [Google Scholar]
- 73. Daya-Grosjean L. and Sarasin A (2005) The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skin tumors. Mutat. Res., 571, 43–56. [DOI] [PubMed] [Google Scholar]
- 74. Stracker T. H. and Petrini J. H (2011) The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol., 12, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee-Theilen M., Matthews A. J., Kelly D., Zheng S. and Chaudhuri J (2011) CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol., 18, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Heyer W. D., Ehmsen K. T. and Liu J (2010) Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet., 44, 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Antoniou A. C., Sinilnikova O. M., Simard J., et al. ; Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers Study (GEMO); Epidemiological Study of BRCA1 and BRCA2 Mutation Carriers (EMBRACE); German Consortium for Hereditary Breast and Ovarian Cancer (GCHBOC); Kathleen Cuningham Consortium for Research into Familial Breast Cancer (kConFab); Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) (2007) RAD51 135G–>C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am. J. Hum. Genet., 81, 1186–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kuchenbaecker K. B., Hopper J. L., Barnes D. R., et al. ; BRCA1 and BRCA2 Cohort Consortium (2017) Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA, 317, 2402–2416. [DOI] [PubMed] [Google Scholar]
- 79. Tutt A. N. J., Lord C. J., Mccabe N., et al. (2005) Exploiting the DNA repair defect in brca mutant cells in the design of new therapeutic strategies for Cancer. Cold Spring Harb. Symp. Quant. Biol., 70, 139–148. [DOI] [PubMed] [Google Scholar]
- 80. Strickland K. C., Howitt B. E., Shukla S. A., et al. (2016) Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget, 7, 13587–13598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mateos-Gomez P. A., Gong F., Nair N., Miller K. M., Lazzerini-Denchi E. and Sfeir A (2015) Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature, 518, 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ceccaldi R., Liu J. C., Amunugama R., et al. (2015) Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature, 518, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Acharya S., Wilson T., Gradia S., Kane M. F., Guerrette S., Marsischky G. T., Kolodner R. and Fishel R (1996) hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U. S. A., 93, 13629–13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hsieh P. and Zhang Y (2017) The Devil is in the details for DNA mismatch repair. Proc. Natl. Acad. Sci. U. S. A., 114, 3552–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wilson T., Guerrette S. and Fishel R (1999) Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J. Biol. Chem., 274, 21659–21664. [DOI] [PubMed] [Google Scholar]
- 86. Palombo F., Iaccarino I., Nakajima E., Ikejima M., Shimada T. and Jiricny J (1996) hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol., 6, 1181–1184. [DOI] [PubMed] [Google Scholar]
- 87. Fishel R., Lescoe M. K., Rao M. R., Copeland N. G., Jenkins N. A., Garber J., Kane M. and Kolodner R (1993) The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell, 75, 1027–1038. [DOI] [PubMed] [Google Scholar]
- 88. Sehgal R., Sheahan K., O’Connell P. R., Hanly A. M., Martin S. T. and Winter D. C (2014) Lynch syndrome: an updated review. Genes (Basel)., 5, 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mensenkamp A. R., Vogelaar I. P., van Zelst-Stams W. A., et al. (2014) Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology, 146, 643–646.e8. [DOI] [PubMed] [Google Scholar]
- 90. Adam R., Spier I., Zhao B., et al. (2016) Exome sequencing identifies biallelic MSH3 germline mutations as a recessive subtype of colorectal adenomatous polyposis. Am. J. Hum. Genet., 99, 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Germano G., Lamba S., Rospo G., et al. (2017) Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature, 552, 116–120. [DOI] [PubMed] [Google Scholar]
- 92. Neefjes J. and Ovaa H (2013) A peptide’s perspective on antigen presentation to the immune system. Nat. Chem. Biol., 9, 769–775. [DOI] [PubMed] [Google Scholar]
- 93. Backert L. and Kohlbacher O (2015) Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med., 7, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ebrahimi-Nik H., Bassani-Sternberg M. and Srivastava P. K (2019) Mass spectrometry-driven exploration reveals nuances of neoepitope-driven tumor rejection. JCI Insight, 4, e129152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B. and Nielsen M (2017) NetMHCpan-4.0: improved Peptide-MHC Class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol., 199, 3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bassani-Sternberg M., Bräunlein E., Klar R., et al. (2016) Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun., 7, 13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ostroumov D., Fekete-Drimusz N., Saborowski M., Kühnel F. and Woller N (2018) CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell. Mol. Life Sci., 75, 689–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mouw K. W., Goldberg M. S., Konstantinopoulos P. A. and D’Andrea A. D (2017) DNA damage and repair biomarkers of immunotherapy response. Cancer Discov., 7, 675–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Le D. T., Durham J. N., Smith K. N., et al. (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science, 357, 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. McGranahan N., Furness A. J., Rosenthal R., et al. (2016) Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science, 351, 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Anagnostou V., Smith K. N., Forde P. M., et al. (2017) Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov., 7, 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kreiter S., Vormehr M., van de Roemer N., et al. (2015) Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature, 520, 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Saloura V., Cohen E. E., Licitra L., Billan S., Dinis J., Lisby S. and Gauler T. C (2014) An open-label single-arm, phase II trial of zalutumumab, a human monoclonal anti-EGFR antibody, in patients with platinum-refractory squamous cell carcinoma of the head and neck. Cancer Chemother. Pharmacol., 73, 1227–1239. [DOI] [PubMed] [Google Scholar]
- 104. Pujade-Lauraine E., Ledermann J. A., Selle F., et al. (2017) Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. The lancet oncology, 18, 1274–1284. [DOI] [PubMed] [Google Scholar]
- 105. Poole, R. M. (2014). Pembrolizumab: first global approval. Drugs, 74, 1973–1981. [DOI] [PubMed] [Google Scholar]
- 106. Agarwala S. S. and Kirkwood J. M (2000) Temozolomide, a novel alkylating agent with activity in the central nervous system, may improve the treatment of advanced metastatic melanoma. Oncologist, 5, 144–151. [DOI] [PubMed] [Google Scholar]
- 107. Hwu W., Ivan D., Prieto V. G., et al. (2008). Randomized phase II neoadjuvant study of temozolomide (TMZ) alone or with pegylated interferon-alfa 2b (PGI) in patients with resectable AJCC stage IIIC or stage IV (M1a) metastatic melanoma. Journal of Clinical Oncology, 26, 20024. [Google Scholar]
- 108. Xie C., Duffy A. G., Mabry-Hrones D., et al. (2019) Tremelimumab in combination with microwave ablation in patients with refractory biliary tract cancer. Hepatology, 69, 2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. A Trial of Radiotherapy and Durvalumab in DLBCL - Full Text View - ClinicalTrials.gov identifier: NCT03610061.
- 110. Wang Y., Deng W., Li N., Neri S., Sharma A., Jiang W. and Lin S. H (2018) Combining immunotherapy and radiotherapy for cancer treatment: current challenges and future directions. Front. Pharmacol., 9, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ventriglia J., Paciolla I., Pisano C., et al. (2017) Immunotherapy in ovarian, endometrial and cervical cancer: state of the art and future perspectives. Cancer Treat. Rev., 59, 109–116. [DOI] [PubMed] [Google Scholar]
- 112. Goh G., Walradt T., Markarov V., et al. (2016) Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget, 7, 3403–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Balachandran V. P., Łuksza M., Zhao J. N., et al. ; Australian Pancreatic Cancer Genome Initiative; Garvan Institute of Medical Research; Prince of Wales Hospital; Royal North Shore Hospital; University of Glasgow; St Vincent’s Hospital; QIMR Berghofer Medical Research Institute; University of Melbourne, Centre for Cancer Research; University of Queensland, Institute for Molecular Bioscience; Bankstown Hospital; Liverpool Hospital; Royal Prince Alfred Hospital, Chris O’Brien Lifehouse; Westmead Hospital; Fremantle Hospital; St John of God Healthcare; Royal Adelaide Hospital; Flinders Medical Centre; Envoi Pathology; Princess Alexandria Hospital; Austin Hospital; Johns Hopkins Medical Institutes; ARC-Net Centre for Applied Research on Cancer (2017) Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature, 551, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Łuksza M., Riaz N., Makarov V., et al. (2017) A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature, 551, 517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Knijnenburg T. A., Wang L., Zimmermann M. T., et al. ; Cancer Genome Atlas Research Network (2018) Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep., 23, 239–254.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]