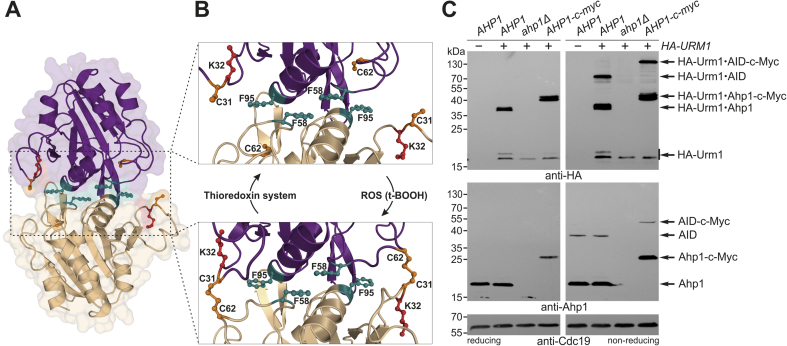
Fig. 1.
Ahp1 structure, redox states and in vivo urmylation. (A) Overview of the Ahp1 homodimer (PDB #4DSR, 4DSQ) composed of two subunits (magenta & beige). Highlighted are residues critical for dimerization (F58 & F95: teal), peroxidase activity (C31 & C62: orange) or known urmylation (K32: red). (B) The enlargement (top panel) shows the redox-active centers formed between each subunit by resolving (C31) and peroxidatic (C62) thiols. Upon oxidation by ROS (t-BOOH), they become disulfide-bridged (bottom panel) and can be reduced by the thioredoxin system (see Fig. 2A). (C) Formation of HA-Urm1•Ahp1 conjugates in vivo. Shown are EMSAs under reducing (left panels) and non-reducing (right panels) conditions on protein extracts from indicated strains expressing HA-URM1 (+) or not (−). NEM-stabilized urmylation was studied by anti-HA blots (top panels) diagnostic for free HA-Urm1 (~17 kDa) and urmylated forms of Ahp1 (~36 kDa) or Ahp1 intersubunit disulfide (AID ~72 kDa) as well as Urm1-modified c-Myc tagged Ahp1 (~43 kDa) or AID (~90 kDa) forms. anti-Ahp1 Western blots (middle panels) detect unmodified Ahp1 (~19 kDa) and AID (~38 kDa) or c-Myc tagged Ahp1 (~27 kDa) and AID (~54 kDa). Protein loading control used anti-Cdc19 blots (bottom panels). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)