Figure 5.
Global Transcriptomic Analysis for SMC4PTD in Activated Gametocytes by RNA-Seq
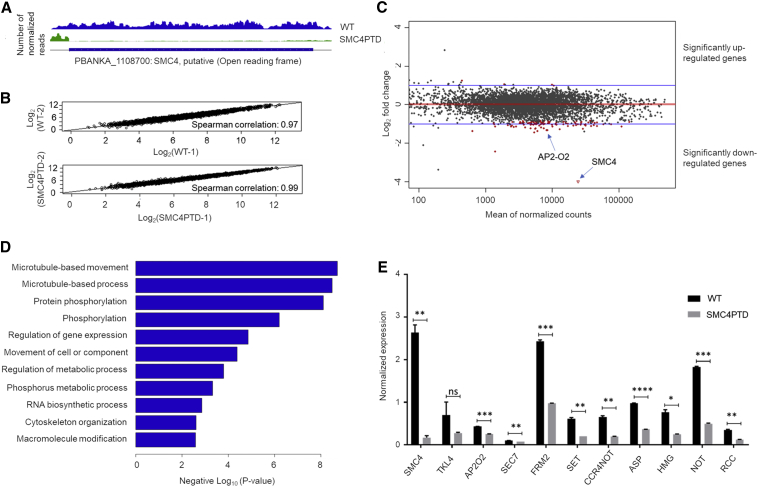
(A) Confirmation of successful depletion of SMC4 transcript in the SMC4PTD line. Tracks shown each represent the average of two biological replicates.
(B) Log-normalized scatterplots demonstrating high correlation between replicates for genome-wide expression. The Spearman correlation was calculated using read counts normalized by number of mapped reads per million.
(C) MA plot summarizing RNA-seq results. M, log ratio; A, mean average. Every gene is placed according to its log fold expression change in the SMC4PTD line compared with the WT line (y axis), and average expression level across replicates of both lines (x axis). Red indicates statistical significance of differential expression at the false-positive threshold of 0.05. 105 genes are downregulated in the SMC4PTD line, and 5 genes are upregulated.
(D) GO enrichment analysis of genes with log10 fold expression change of −0.5 or lower in the SMC4PTD line.
(E) qRT-PCR analysis of selected genes identified as downregulated in (C), comparing transcript levels in WT and SMC4PTD samples. Error bar, ± SEM, n = 3.
Unpaired t test was performed for statistical analysis: ∗p < 0.05 ∗∗p < 0.001, ∗∗∗p < 0.0001, and ∗∗∗∗p < 0.00001. See also Figure S5 and Tables S3 and S4.