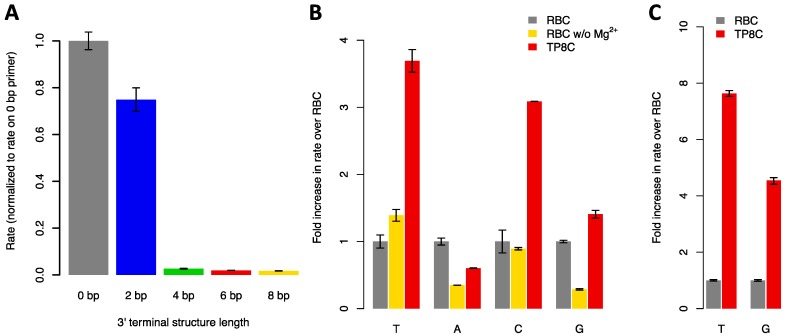
Figure 1.
(A) Elongation of DNA primers with 3′ terminal hairpins of varying length with ddTTP using MTdTwt in RBC reaction buffer (50 mM potassium acetate, 20 mM Tris acetate, 10 mM magnesium acetate, 0.25 mM cobalt chloride, pH 7.9) at 37 °C (n = 3 replicates). Elongation rates were normalized to rates on the unstructured substrate (0 bp). (B) Elongation of an unstructured primer (P1) with ddNTPs using MTdTwt in reaction buffer (50 mM potassium acetate, 20 mM Tris acetate, pH 7.9) with varying M2+ concentrations: RBC (10 mM Mg2+, 0.25 mM Co2+), RBC without Mg2+ (0.25 mM Co2+), and TP8C (1 mM Co2+) (n = 2 replicates). Elongation rates were normalized to rates in RBC. (C) Elongation of an 8 bp hairpin primer (P5) with ddTTP or ddGTP in RBC and TP8C using MTdTwt (n = 3 replicates). Elongation rates were normalized to rates in RBC. In all of the above, error bars correspond to mean ± SD; T = ddTTP; A = ddATP; C = ddCTP; G = ddGTP.