Figure 3.
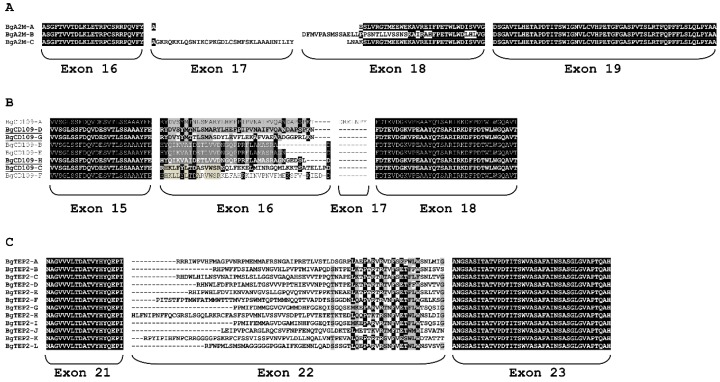
ClustalW alignment of alternative splice variants for three BgTEP proteins. Amino acids encoded by alternative exons corresponding to the highly variable region of some BgTEP were aligned, together with amino acids from border conserved exons, using clustalW. Identical amino acids are highlighted in black, similar ones in grey. (A) Alignment of BgA2M variants showing alternative splicing in exons 17 and 18. (B) Alignment of BgCD109 splice variants. Underlined variant names correspond to variants recovered from Biomphalaria glabrata transcriptomes. The other variants were only predicted from BglaB1 genome assembly. (C) Alignment of the 12 BgTEP2 splice variants.