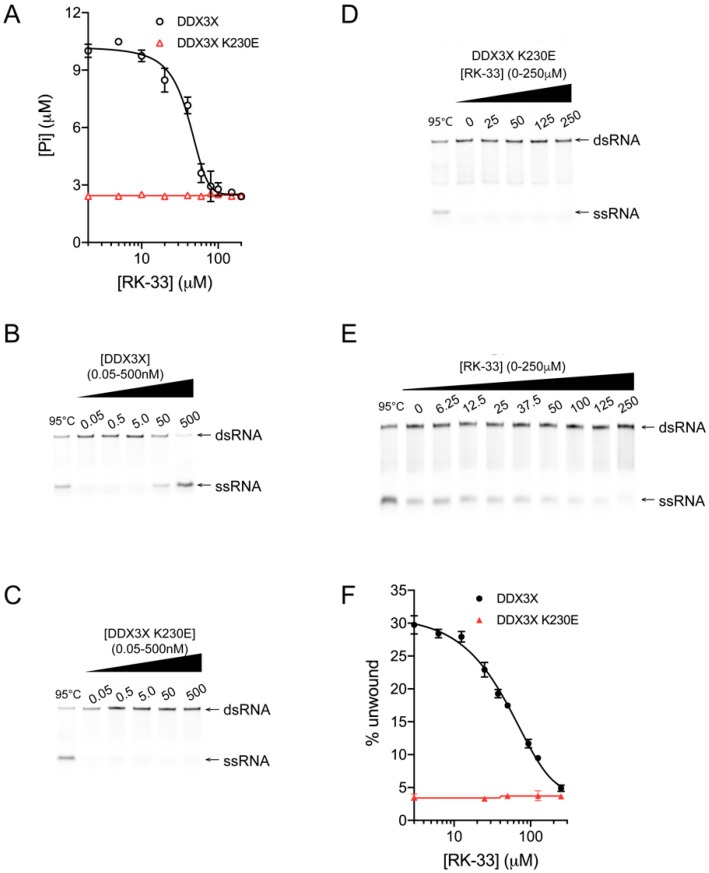
Figure 2.
RK-33 inhibits DDX3X helicase and ATPase activity. DDX3X (black) and DDX3X K230E (red) proteins were assessed for their ability to (A) hydrolyze ATP and (B–F) unwind a synthetic analogue of duplex RNA (poly(I:C)), in the presence of increasing concentrations of RK-33 or equivalent % DMSO. (A) The effect of up to 200 μM RK-33 on the formation of free inorganic phosphate (Pi) by DDX3X (IC50 = 40 μM) was determined by a colorimetric assay, where free phosphate reacts with the Biomol® Green reagent to initiate color development quantified at 620 nm. Results represent the mean +/− standard error of the mean (SEM) for pentaplicate wells from a single assay, representative of two (n = 2) independent experiments; (B,C) The capacity of DDX3X and DDX3X K230E to unwind Cy5-labeled double-stranded RNA (dsRNA) into single-stranded RNA (ssRNA) was determined by a helicase assay. Following incubation of 0–0.5 μM DDX3X or DDX3X K230E with dsRNA for 30 min at 37 °C, reactions were stopped by the addition of STOP solution (0.6% SDS, 60 mM EDTA, 40% (w/v) sucrose and 0.25% bromophenol blue). Cy5-labeled dsRNA and ssRNA were visualized by fluorescence on polyacrylamide gel. (D–F) The effect of RK-33 or equivalent % DMSO on the capacity of DDX3X to unwind dsRNA (IC50 = 35 μM) was determined. Cy5-labeled dsRNA and ssRNA were visualized by fluorescence on polyacrylamide gels in (D) and (E) were quantified using Image Studio Lite, with the ratio of the intensities for the ssRNA:dsRNA bands plotted as a % unwound in (F). Results in (F) represent the mean ± SEM for three independent experiments (n = 3)—of which, (D) and (E) are representative. A non-linear, sigmoidal dose response model was fit to the RK-33 concentration-activity data using GraphPad Prism 8 to generate figures. Lane 1 in each gel depicts heat-denatured RNA.