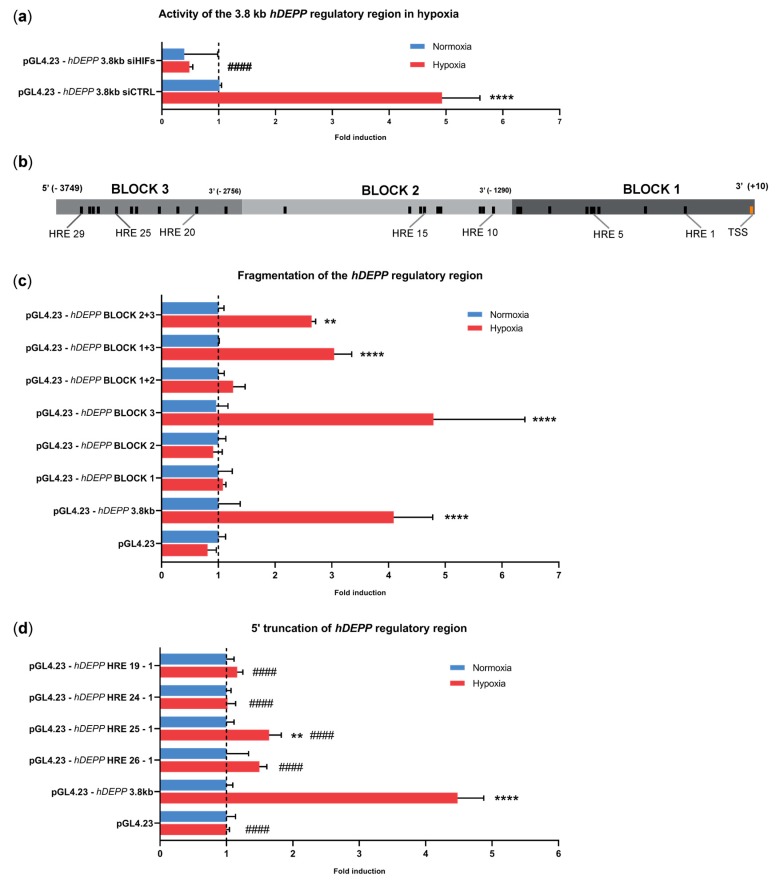
Figure 3.
Hypoxic regulation by the 3.8 kb DNA fragment upstream of the DEPP transcriptional start site. (a) Luciferase activity driven by the 3.8 kb hDEPP regulatory region assayed in hypoxic and normoxic ARPE-19 cells. Cells were transfected with the reporter plasmid containing DEPP regulatory sequence and treated with scrambled siRNA (siCTRL) or with siRNAs against HIF1A and HIF2A (siHIFs). Luciferase activity was expressed relatively to the activity in normoxic ARPE-19 cells treated with control siRNA (pGL4.23-hDEPP 3.8 kb siCTRL). (b) Schematic representation of the 3.8 kb DNA fragment isolated from ARPE-19 genomic DNA and cloned into the luciferase reporter vector pGL4.23. Putative HREs are shown as black bars. The HRE closest to the TSS is numbered as HRE 1 and the most distal one as HRE 29. Precise positions of the HREs are given in Table S1. The DNA fragment was subdivided into three BLOCKS, as indicated. (c) Luciferase activity in cells transfected with the indicated BLOCK constructs was expressed relative to the respective normoxic controls, which were set to 1. (d) Luciferase activity in cells transfected with 5′ truncated constructs. HRE numbers in the construct name indicate the presence of the respective HRE sequences. Luciferase activities were expressed relative to the respective normoxic controls, which were set to 1. All luciferase values were normalized to b-galactosidase activity from the co-transfected normalizer plasmid. The empty pGL4.23 plasmid served as negative control. HRE: hypoxia responsive element. Shown are means ± SD of N = 3. Statistics: One-way ANOVA with the Brown–Forsythe test, *: p ≤ 0.05, **: p ≤ 0.005, ***: p ≤ 0.005, ****: p ≤ 0.0005, ####: p ≤ 0.0005. Stars (*) denote significance between the hypoxic and normoxic activity of the indicated vector. Hashtags (#) indicate significance between the hypoxic activity of the indicated vector and hypoxic activity of the reporter vector containing the 3.8 kb regulatory region (pGL4.23-hDEPP3.8 kb).