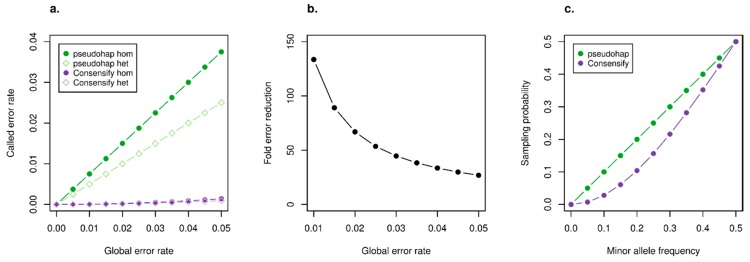
Figure 1.
Expected performance of Consensify compared with standard pseudohaploidisation, assuming equal base composition and equal error probabilities across all nucleotides. (a) Shows the expected called error rates (y axis) across a range of global error rates (x axis) for standard pseudohaploidisation (green) and Consensify (purple), for both homozygous sites (circles) and heterozygous sites (rhombuses). (b) Shows the fold-reduction in error rates achieved by using Consensify compared with pseudohaploidisation (y axis), for a range of global error rates (x axis). Note that the fold-reduction in error is equal for both homozygous and heterozygous sites. (c) Shows the probability of sampling (y axis) an allele which is underrepresented in the read stack (x axis) using Consensify and standard pseudohaploidisation.