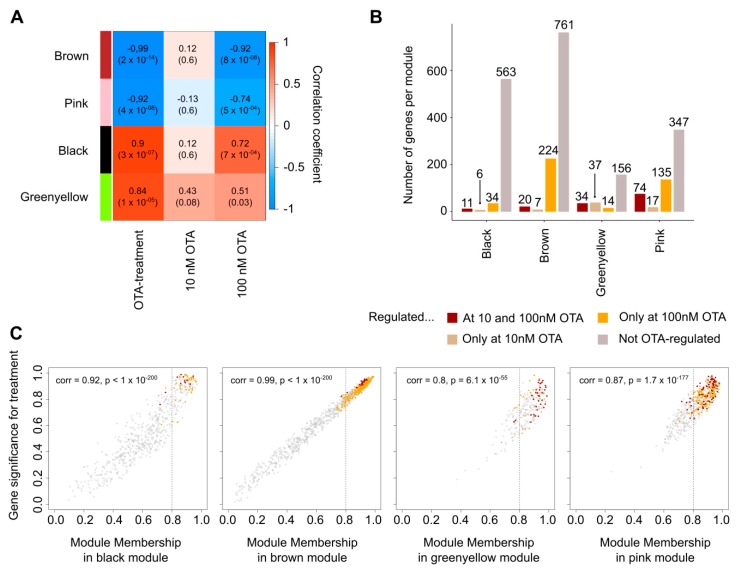
Figure 3.
Weighted correlation network analysis was used to identify groups of genes putatively involved in OTA-induced cell-cycle dysregulation. (A) The heatmap represents the modules with high correlation between their Eigengenes and the “OTA-treatment” trait (abs(corr) ≥ 0.75) and with abs(corrtreatment) > abs(corr100nM OTA) (to avoid unspecific effects due to too high OTA concentrations). (B) The numbers of genes found to be regulated by OTA (by differential expression analyses) in modules from (A) are displayed. No distinction between the cell types is made in this figure (overlap of the analyses output). (C) The gene significance for the trait “OTA treatment” and the module membership were calculated for each of the modules presented in (A). Each dot corresponds to a gene. The dashed vertical line corresponds to the threshold (0.8) used for selecting hub-like genes. See also Figures S6 and S7 (Supplementary Materials).