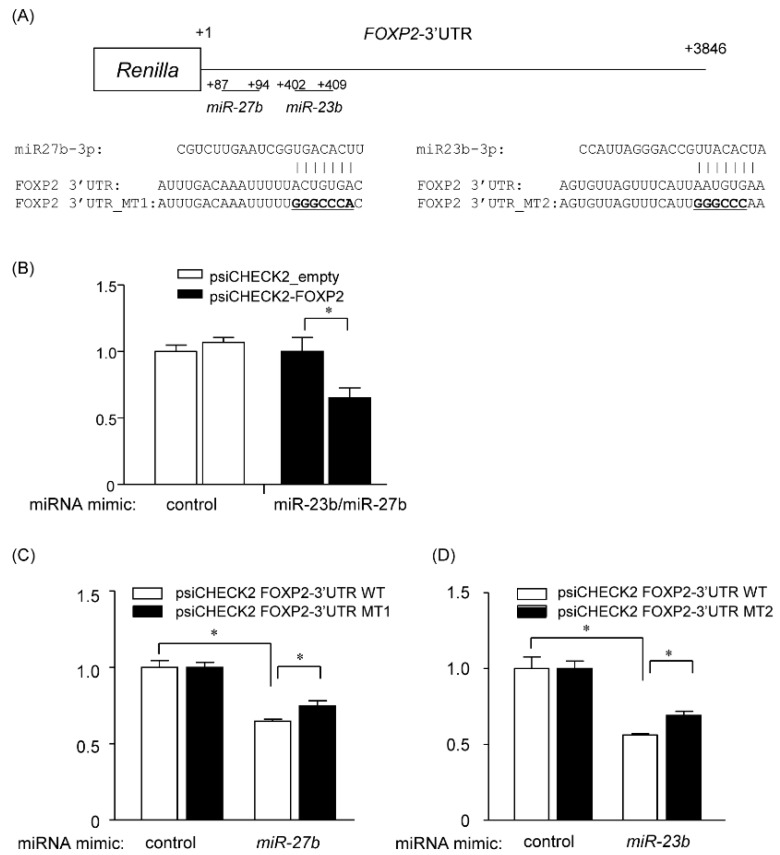
Figure 7.
FOXP2 is directly targeted by both miR-23b and miR-27b. (A) miRDB predicted a putative binding site of miR-23b (nt 402–409) and of miR-27b (nt 87–94) in the FOXP2 3’ UTR region. The mutated (MT) sequences of the putative binding sites are shown as FOXP2 3′UTR_MT1 or MT2. (B) psiCHECK2 vector containing the full length of 3′ UTR in FOXP2 or empty vector was co-transfected with both miR-23b and miR-27b. Renilla luciferase activity was normalized to firefly luciferase. Fold-change values were normalized to empty vector. (C,D) psiCHECK2 vector containing the full length of 3′ UTR in FOXP2 (psiCHECK2 FOXP2-3′UTR WT) or the mutated binding sequences (psiCHECK2 FOXP2-3′UTR MT1 or MT2) were co-transfected with either mimic miR-23b or miR-27b. Renilla luciferase activity was normalized to firefly luciferase. Fold-change values were normalized to empty vector. Fold-change values were then normalized to mimic control-treated cells. Data are expressed as the mean ± standard deviation (SD; n = 4). * Statistically significant difference versus control (B–D) or WT (C,D) (unpaired Student’s t-test, p < 0.05).