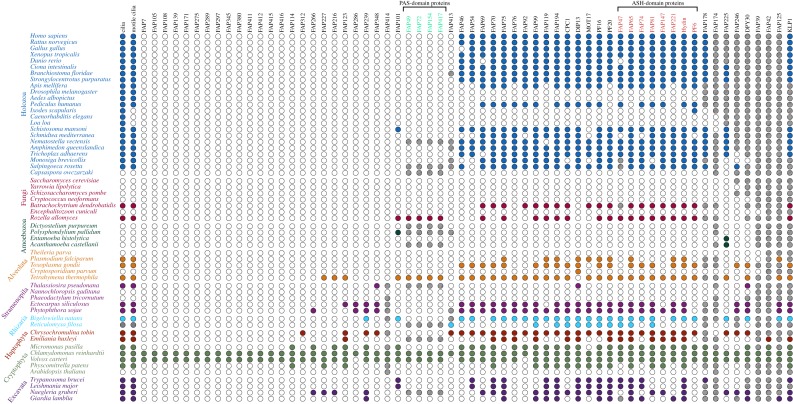
Figure 2.
Dot plot showing phylogenetic distribution of orthologues of C. reinhardtii CA-specific proteins. Species belonging to the same major taxonomic group are indicated by text with the same colour. Coloured dots indicate either the presence of cilia or motile cilia (first and second columns, respectively) or the presence of at least one likely orthologue in the indicated species. Empty dots indicate the absence of cilia, motile cilia or orthologues. Grey dots for Reticulomyxa filosa indicate uncertainty with regard to the presence of cilia (see text). Hatched dots indicate that sequences from these species were present in the OrthoFinder gene tree but eliminated as likely orthologues because they were in a different clade from the C. reinhardtii query protein (see Methods). Proteins most affected in this way were members of large families or contained common conserved motifs and included KLP1 and FAP125 (kinesin superfamily), FAP42 (transferase family), FAP39 (ATPase), DPY30 (DPY-30 domain), FAP246 (LRR repeats and EF-hands), FAP225 (EF-hands), FAP174 (MYC-binding) and FAP178 (calponin domain). PAS- and ASH-domain proteins (green and red font, respectively) are indicated. (Online version in colour.)