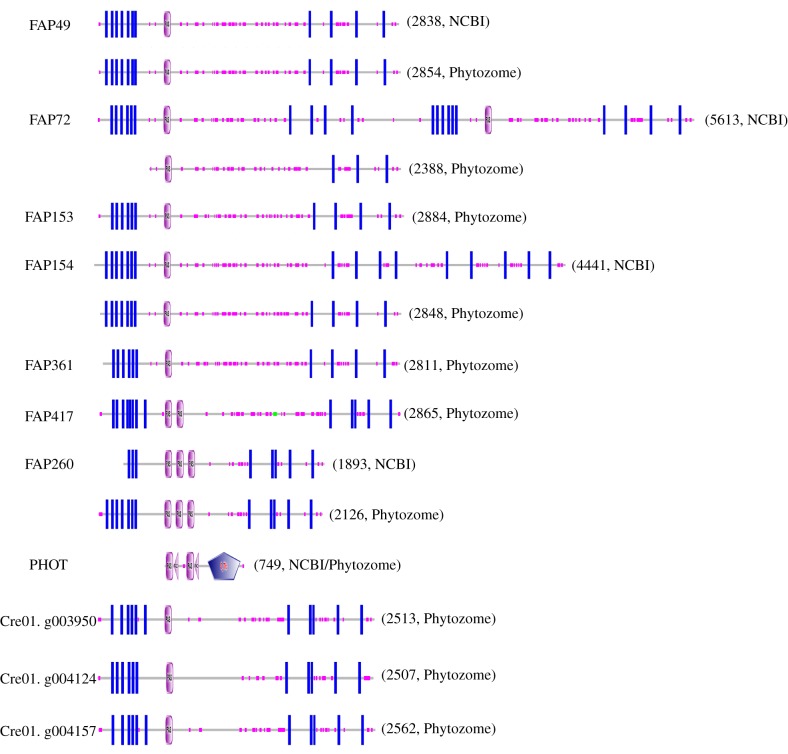
Figure 4.
Domain architecture of predicted C. reinhardtii flagellar proteins containing PAS domains. Pink ovals marked ‘PAS’ indicate predicted PAS domains. Blue rectangles indicate predicted transmembrane domains. Small pink boxes along each protein indicate regions of low sequence complexity. In PHOT, pink triangles marked ‘PAC’ indicate ‘motif C-terminal to PAS motifs (likely to contribute to PAS structural domain)’ and the blue pentagon marked ‘S_TKc’ indicates a ‘serine/threonine protein kinases, catalytic domain’. The number to the right of each architecture indicates the number of amino acids in the protein sequence. PAS domains were identified as described in the Methods. When there is a discrepancy between sequences from NCBI and Phytozome, predictions for both sequences are shown. The locations of exclusive unique peptides suggested that some Phytozome models were more accurate than the corresponding NCBI models; e.g. for FAP72, unique peptides were predicted to originate from throughout the N-terminal half of the NCBI model (most of which is identical to the Phytozome model) but not the C-terminal half. (Online version in colour.)