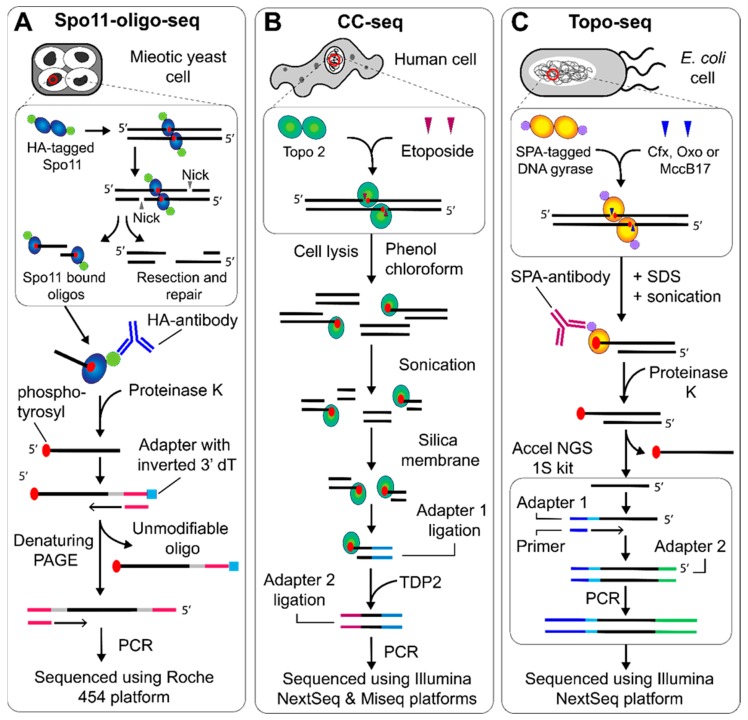
Figure 2.
NGS protocols to map topo cleavage sites genome wide. (A) Spo11-oligo-seq began with DNA extraction from meiotic S. cerevisiae cells. HA-tagged Spo11-bound DNA oligos are enriched with the HA-antibody, before the protein is removed using proteinase K. The 3′ end is extended using TdT and an adapter is ligated that contains an inverted dT residue. The complementary strand is synthesized, followed by separation using denaturing PAGE, 3′ end tailing and adapter ligation on the newly synthesized strand, which can then be sequenced using the Roche 454 platform [12]. (B) CC-seq, used to map topo II cleavage, began with DNA extraction from etoposide treated human cells. The bulk protein is removed using phenol-chloroform extraction, whilst the protein bound to DNA remains in the aqueous phase. The DNA can then be sonicated and enriched using a silica membrane. The first ligation attached adapters to the sonicated end, which was then followed by TDP2-dependent removal of the topo II, before a second adapter ligation followed by sequencing using the Illumina NextSeq and Miseq platforms [15]. (C) Topo-seq, used to map gyrase cleavage, began with DNA extraction from E. coli cells treated with ciprofloxacin (cfx), oxolinic acid (oxo) or microcin B-17 (MccB17), followed by sonication. Gyrase cleavage complexes were then enriched using the SPA-antibody before proteinase K treatment and use in the Accel NGS 1S kit to generate fragments that can be sequenced using the Illumina Nextseq platform [18].