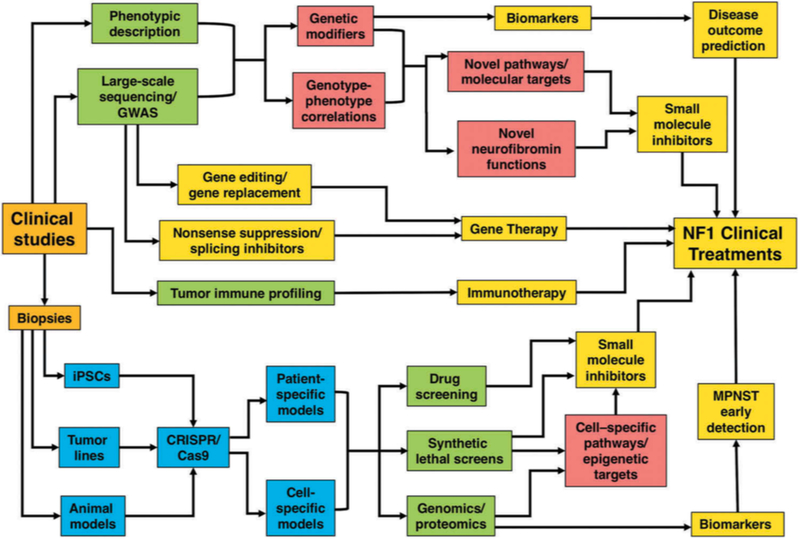
Figure 5. Network of approaches leading to discovery and testing of new therapeutic targets in NF1.
1) Clinical examination of patients combined with molecular analyses is beginning to reveal NF1 genotype-phenotype correlations - findings that may define novel functions of neurofibromin and identify new therapeutic targets. In addition, genome-wide association studies (GWAS) offer a platform to identify genetic modifiers, which will facilitate the identification of novel targets and biomarkers. Understanding the effect of environmental factors and hormones on NF1 disease progression may also reveal novel treatments. 2) Patient-derived biopsies will aid in the generation of: a) induced pluripotent stem cells (iPSCs) to allow development of patient and cell-specific models and give insights for new targets and biomarkers for different NF1 clinical manifestations; b) tumor-specific cell lines and animal models accurately reflecting the human disease that will permit improved screening of small molecule inhibitors; c) patient-derived cell line models will facilitate cellular pathway analysis (in particular RAS pathway and its upstream and downstream effectors (including receptor tyrosine kinases and micro RNAs) identification of therapeutic targets and biomarkers, and will also allow the testing of novel drugs including small molecule inhibitors prior to their use in clinical trials; d) synthetic lethal screening (using CRISPR libraries) could be exploited to devise therapies to selectively kill NF1-deficient tumors; e) immune profiling leading to immunotherapy and generation of novel biomarkers for NF1-associated tumors. 3) Gene therapy approaches focus on antisense oligonucleotides (ASOs) and nonsense suppression, whereas potential correction of mutations via gene editing offers a possibility of restoring endogenous NF1 gene function, thereby providing a long-term solution for NF1 patients. Highlighted boxes: clinical studies/biopsies (orange), genetic analysis and screening (green), disease models (blue), new insights into basic NF1 biology (red), potential therapeutic approaches and clinical treatments (yellow).