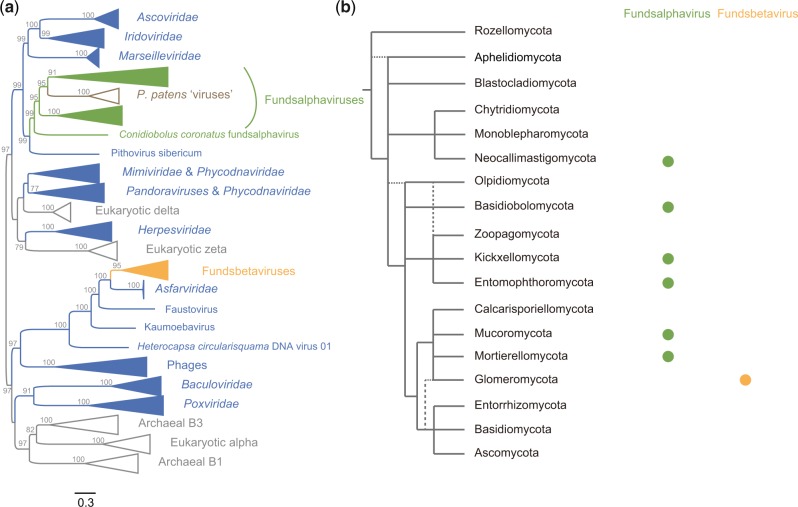
Figure 1.
Phylogeny and host distribution of fungus NCLDV-like sequences. (a) Phylogenetic tree of fundsalphaviruses and fundsbetaviruses based on DNAP. Fundsalphaviruses, fundsbetaviruses, other dsDNA viruses, and P. patens ‘virus’ were labeled in green, orange, blue, and brown, respectively. This unrooted tree was reconstructed using a maximum likelihood method. Ultrafast bootstrap (UFBoot) values (>75) were shown on selected nodes. (b) Host distribution of fundsalphaviruses and fundsbetaviruses. The fungi phylogeny at the level of phylum was inferred from Tedersoo et al. (2018). The presence of fundsalphaviruses and fundsbetaviruses in fungus phylum was labeled with green solid circle and orange solid circle, respectively.