Figure 1.
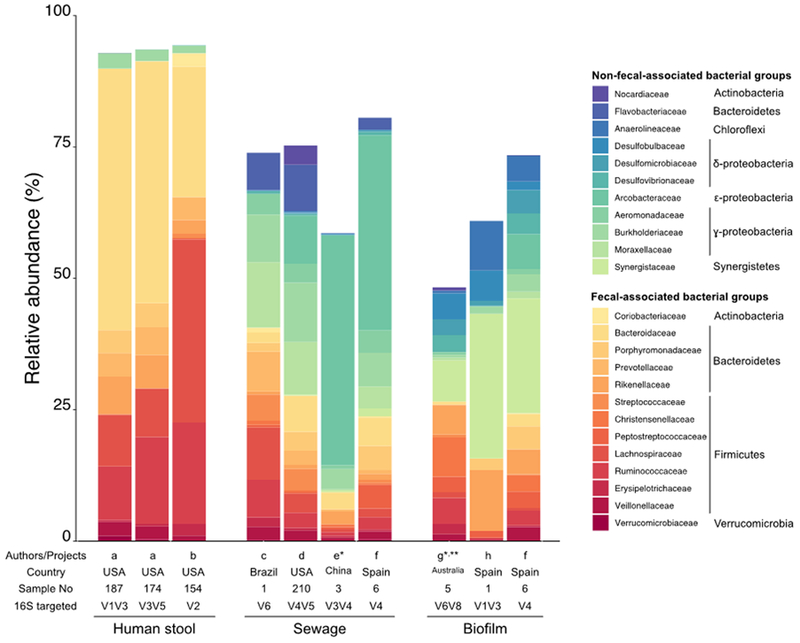
Dominant bacterial families in fecal human microbiome, untreated sewage and sewer biofilm based on 16S rRNA gene amplicon data. Only the top 10 dominant families among at least one of the three environments are displayed. Fecal-derived taxa are represented using red/orange hue colors, while non-fecal-derived taxa are colored in blue/green hue. List of the publications: a [26], b [66], c [67], d [6], e [3], f [19], g [68], h [69]. *: data reprocessed from the SRA files using the 454 Mothur SOP and Silva132 database. **: sewer pilot.
