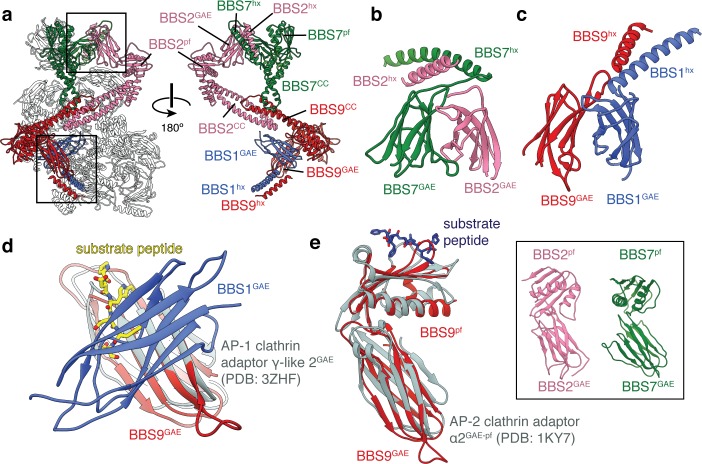
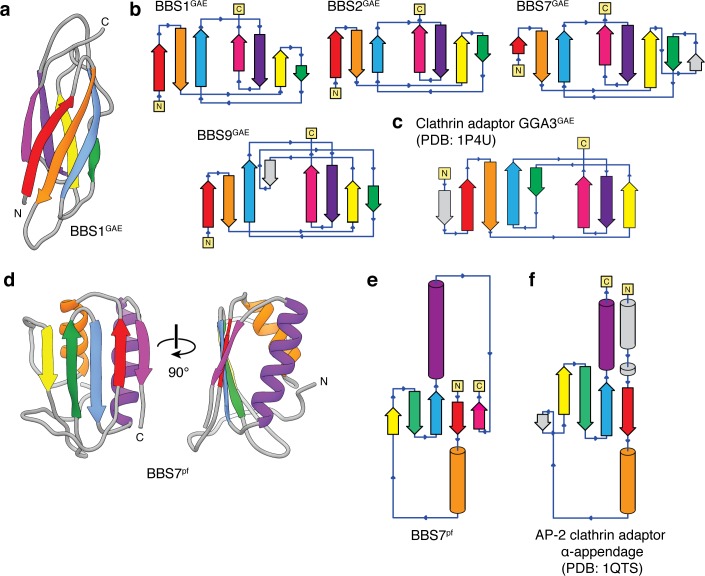
Figure 3. BBS1, BBS2, BBS7 and BBS9 are homologous proteins with similarities to the clathrin adaptor proteins.
(a) Location of BBS1, BBS2, BBS7, and BBS9 in the BBSome, colored except for their β-propeller domains. GAE heterodimers shown in panels c and d are boxed. In the rotated view all non-colored subunits are removed for clarity. (b) Heterodimerization of BBS2 and BBS7 involves the hx-GAE module. (c) Heterodimerization of BBS1 and BBS9. (d) Superposition of BBS9GAE with the GAE domain of AP-1 clathrin adaptor subunit γ-like 2 reveals that the heterodimerization interface with BBS1GAE would occlude the substrate binding pocket. (e) The GAE-pf module of BBS2, BBS7 and BBS9 resembles the equivalent module of the AP-2 clathrin adaptor α2-adaptin. While BBS9GAE-pf superposes closely with α2-adaptin, the GAE and pf domains of BBS2 and BBS7 (inset) adopt different orientations relative to one another.