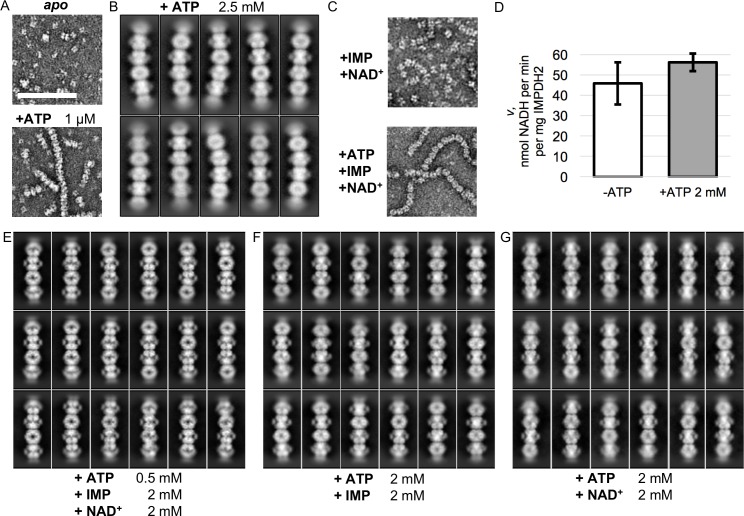
Figure 2. Electron microscopy of uninhibited IMPDH2 filaments.
(A) Negative stain EM of purified human IMPDH2. Treatment with 1 μM ATP induces filament assembly. Scale bar 100 nm. (B) Representative 2D class averages from the 2.5 mM ATP cryo-EM dataset. (C) Negative stain EM of actively catalyzing IMPDH2 (2 mM IMP, 2 mM NAD+), with and without 2 mM ATP. (D) Initial velocity of enzyme (2 mM IMP, 2 mM NAD+), with and without 2 mM ATP. Average of three replicates, error bars + /- 1 s.D. (E–G) Representative 2D class averages from the three uninhibited enzyme cryo-EM datasets, with nucleotide concentrations as indicated at bottom.