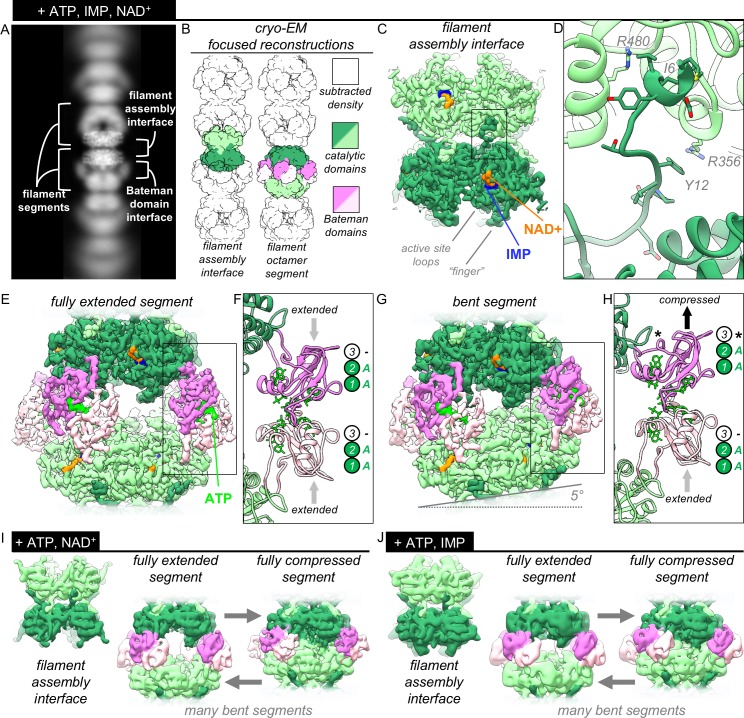
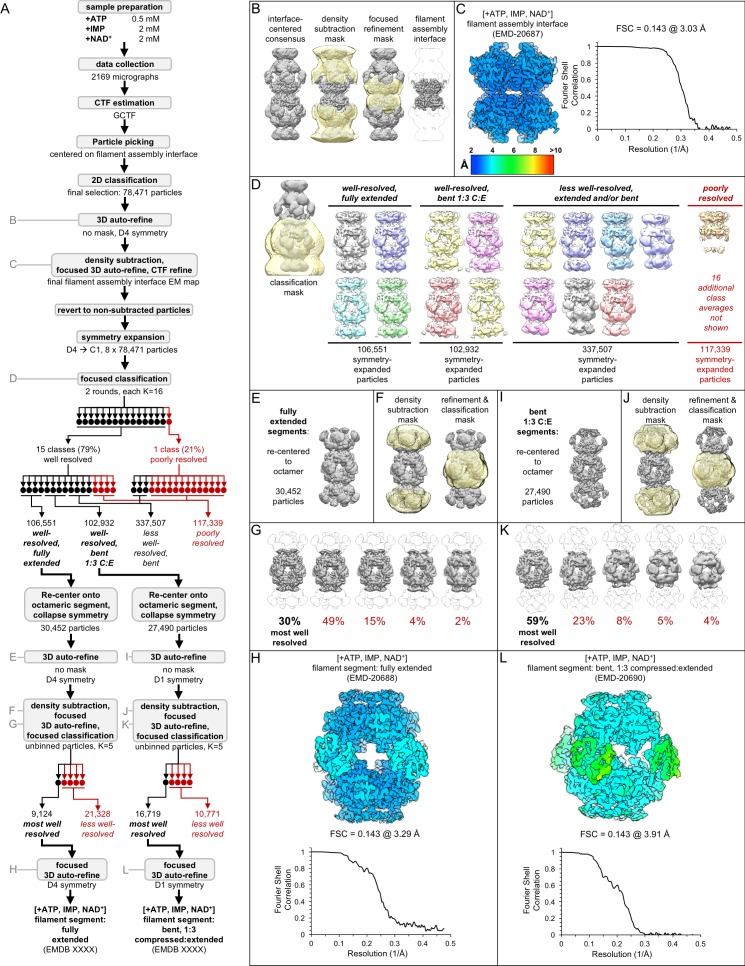
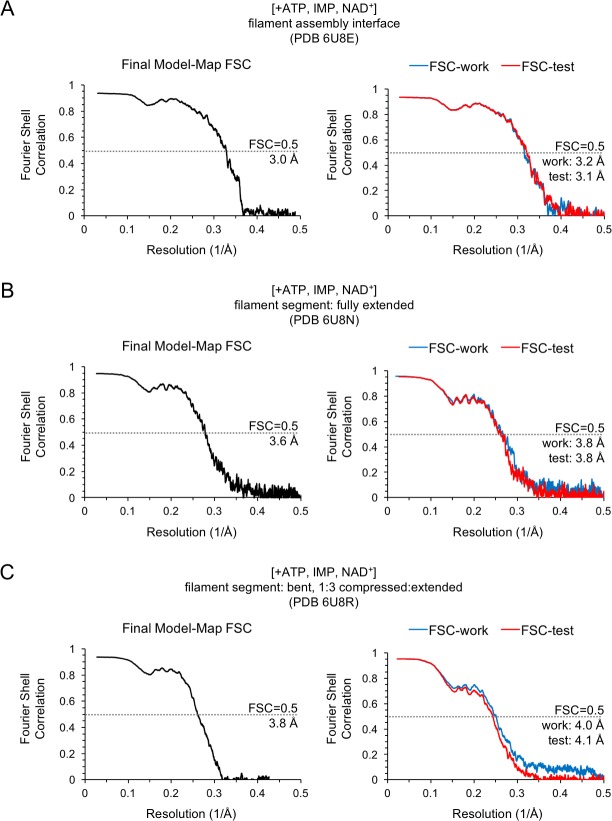
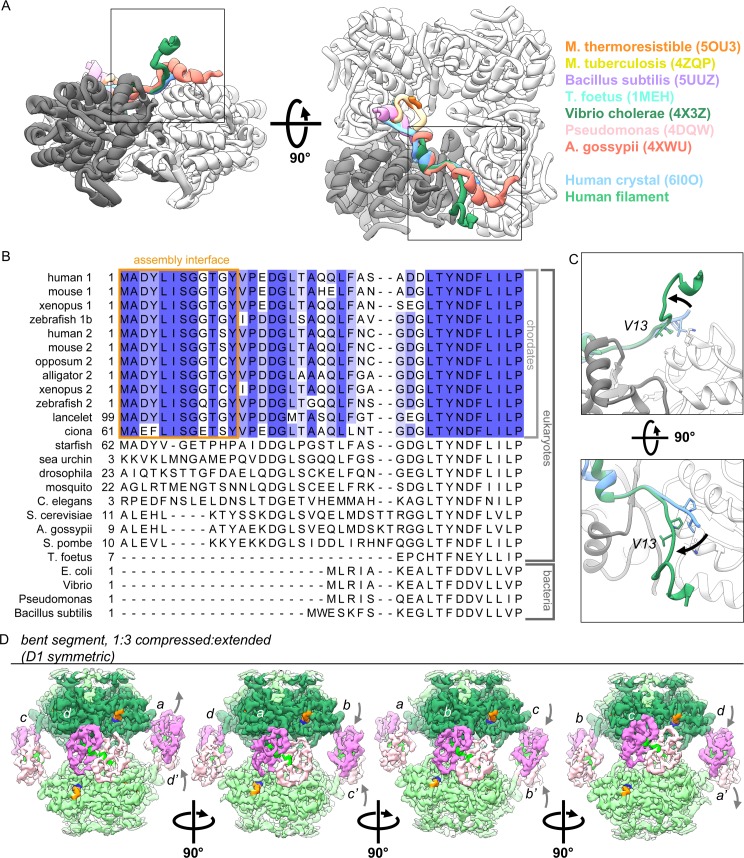
Figure 3. The structures of uninhibited IMPDH2 filaments.
(A) Cryo-EM of IMPDH2 filaments with both substrates (representative 2D class average). (B) We resolved two types of structures from IMPDH filaments: the consensus filament assembly interface, and various conformations of filament segments. (C) Cryo-EM density for the ATP/IMP/NAD+ consensus filament assembly interface, consisting of two tetramers bound back-to-back (dark and light green) (0.5 mM ATP, 2 mM IMP, 2 mM NAD+). (D) The filament assembly interface is mediated by the vertebrate-specific N-terminus, in particular a key bridge between Y12 and R356. (E) Cryo-EM density for the ATP/IMP/NAD+ fully extended filament segment. Opposing catalytic tetramers (dark and light green), are held separate by their symmetrically extended Bateman domains (dark and light pink). ATP (bright green) is resolvable in the Bateman domains. (F) In the fully extended Bateman domains, sites 1 and 2 are occupied by ATP, and site 3 is unformed. (G) Cryo-EM density for the best resolved ATP/IMP/NAD+ bent filament segment, in which the two catalytic tetramers are not parallel. (H) Filament segment bending results from asymmetric compression of Bateman domains. In this reconstruction, one protomer from each of the two tetramers is compressed, and allosteric site 3 is formed, but unoccupied (black asterisk). (I) Summary of the ATP/NAD+ cryo-EM dataset (2 mM ATP, 2 mM NAD+). The filament assembly interface is unchanged, and filament segments varied from fully extended, to bent, to fully compressed. In the absence of IMP, the flexible active site loops are disordered. (J) Summary of the ATP/IMP cryo-EM dataset (2 mM ATP, 3 mM IMP).