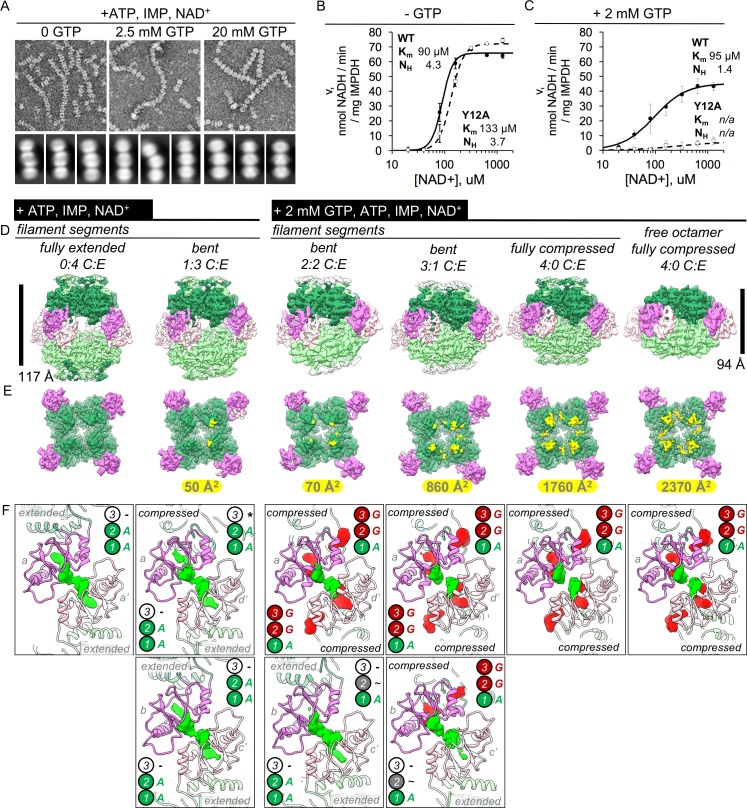
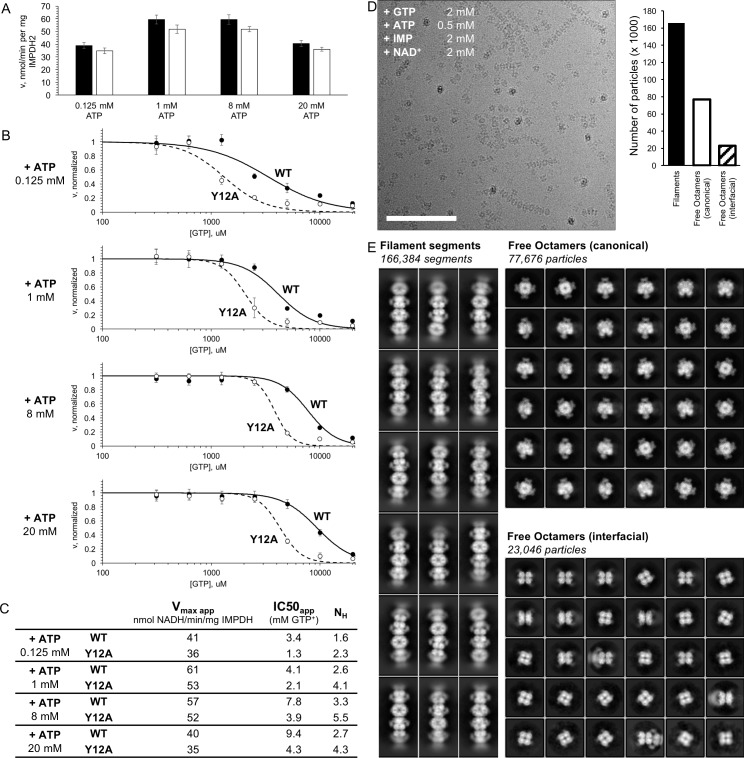
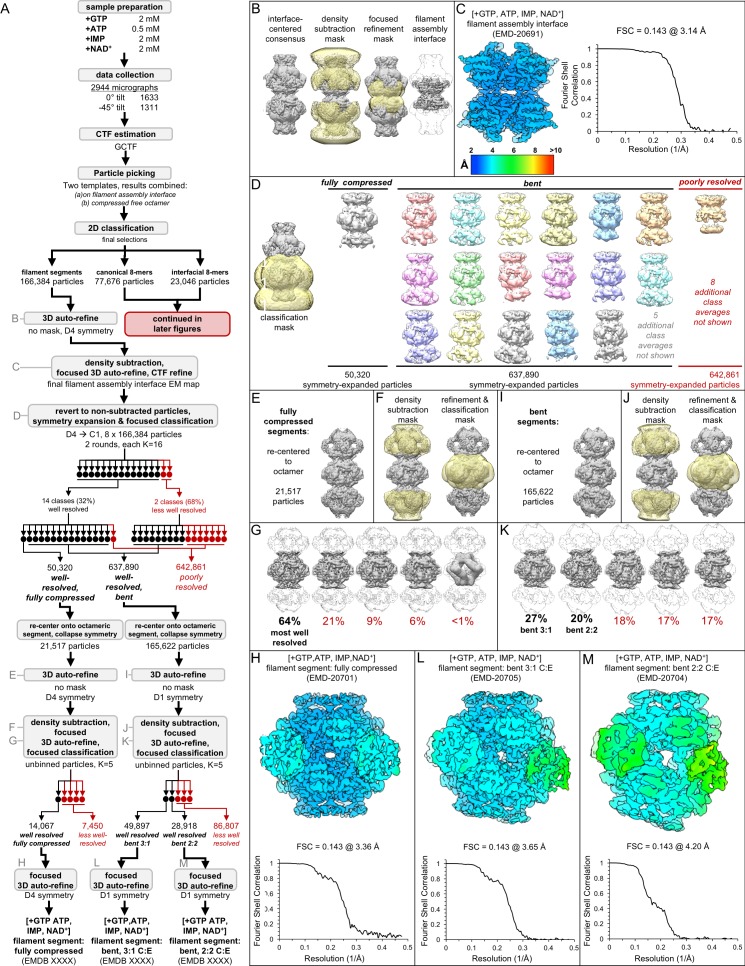
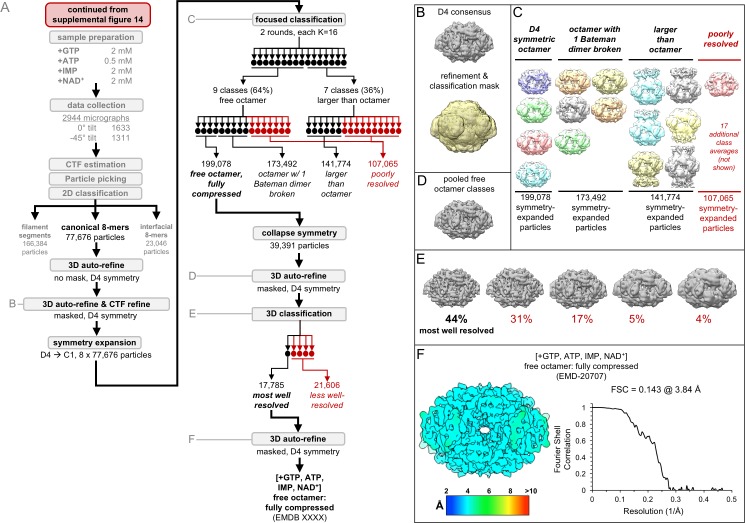
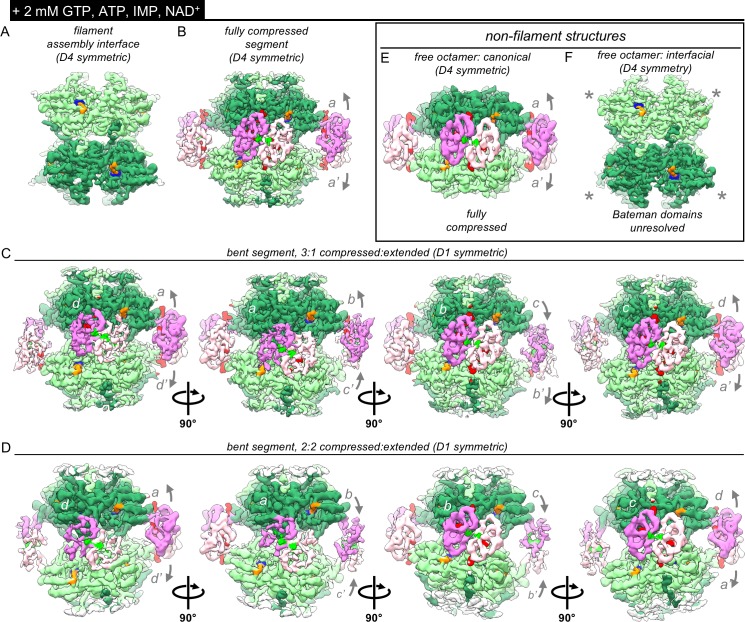
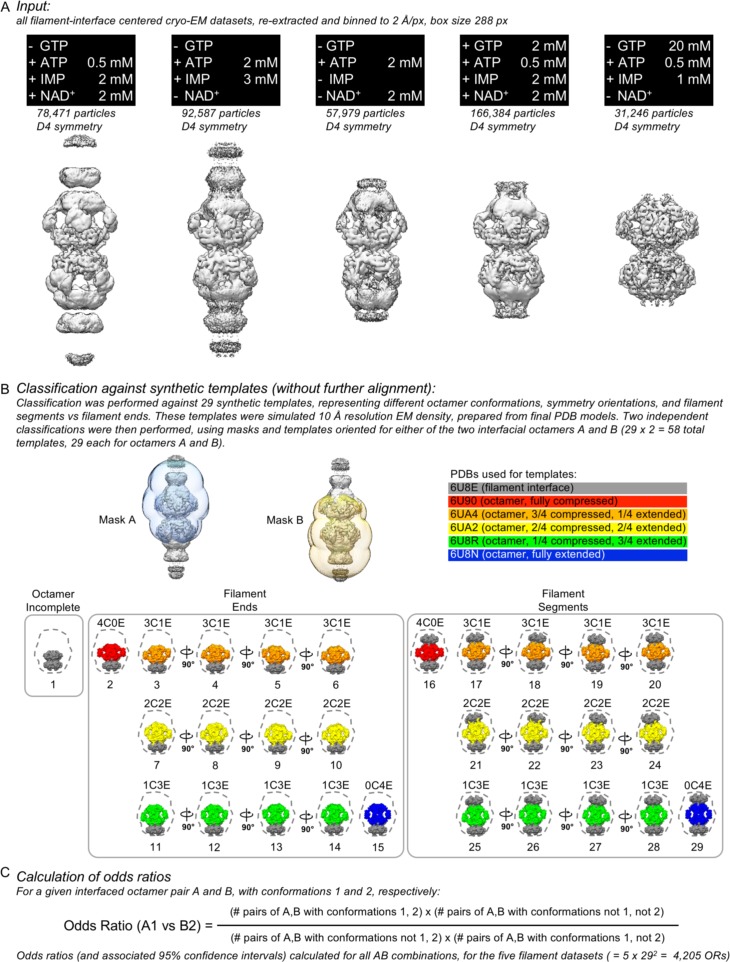
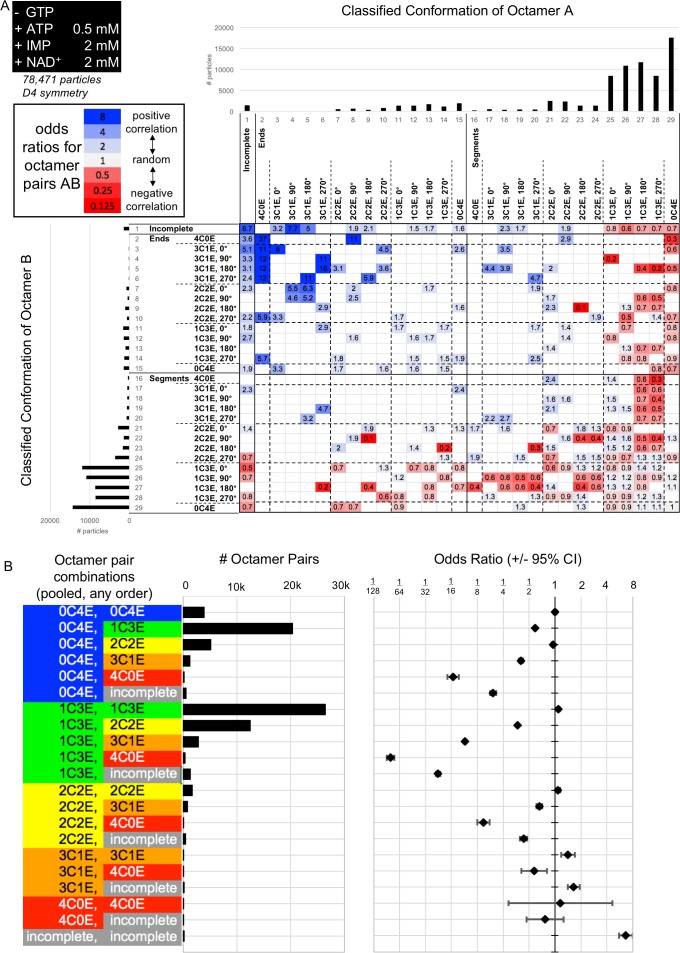
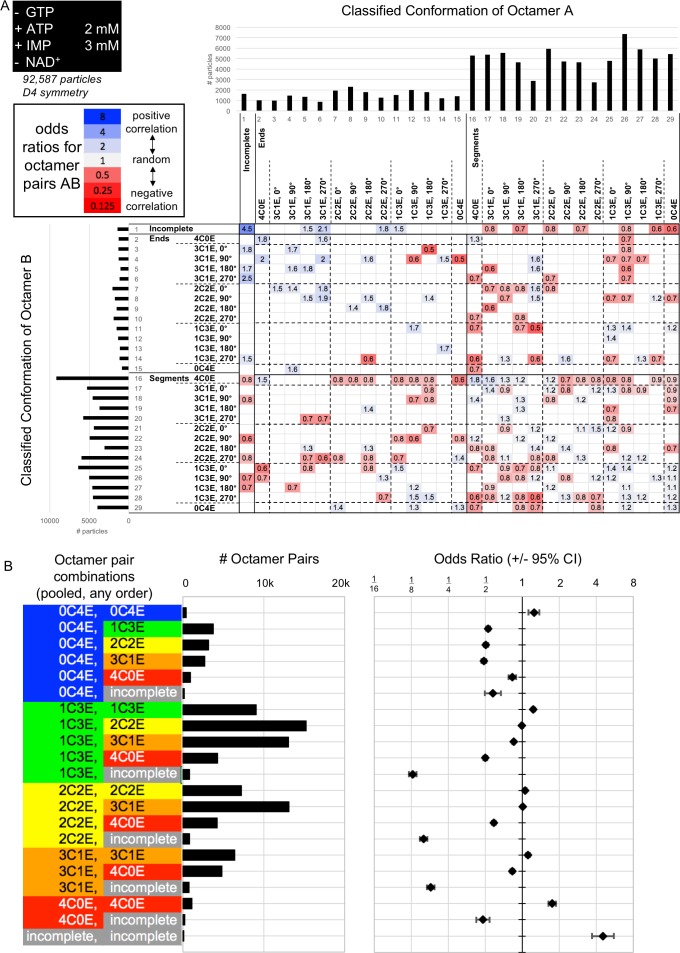
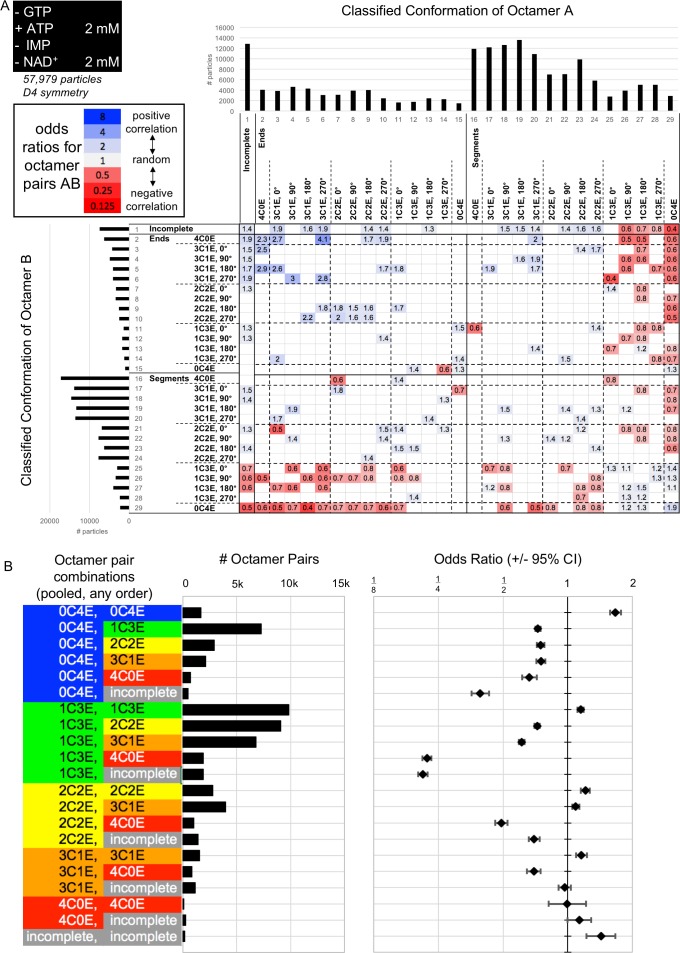
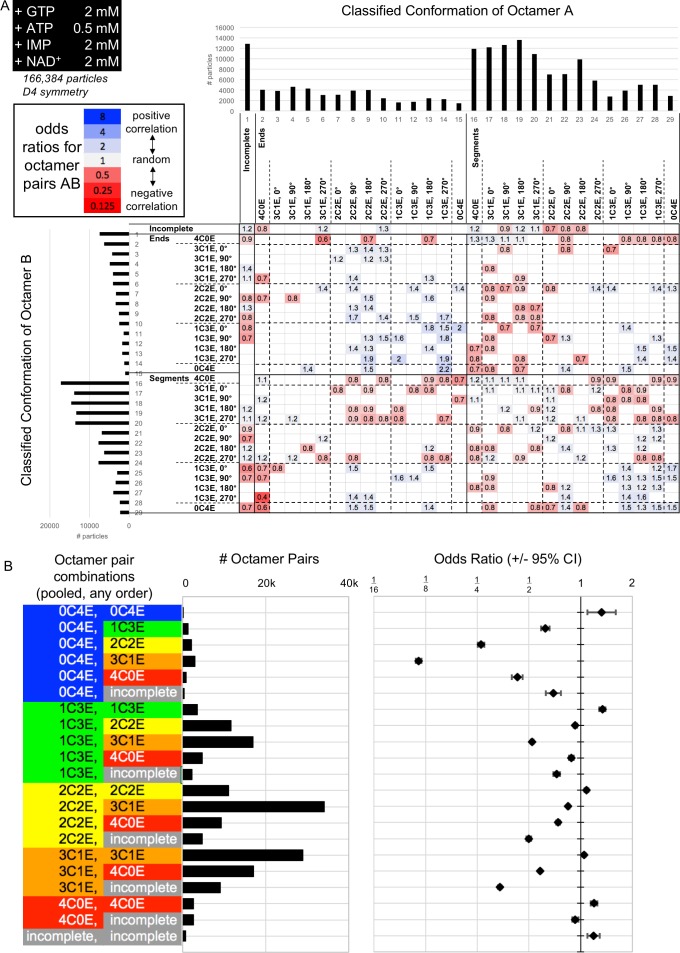
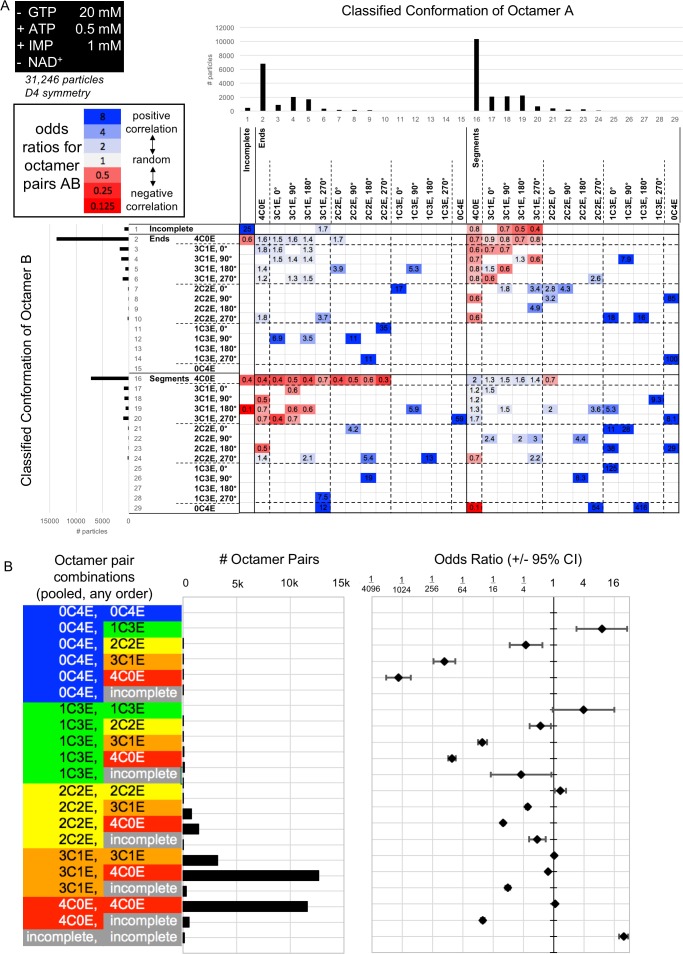
Figure 5. IMPDH2 filaments resist GTP inhibition by promoting bent octamer conformations that separate opposing active sites.
(A) Negatively stained EM of uninhibited (left), partially inhibited (center), and fully inhibited (right) IMPDH2. Representative micrographs and reference free 2D class averages. Prepared with 1 mM IMP, 1 mM NAD+, and either 0, 2.5 mM, or 20 mM GTP. (B–C) NAD+ saturation curves of uninhibited WT IMPDH2 (solid line), and the non-assembly mutant Y12A (dashed line). Reactions performed with 0.5 mM ATP, 1 mM IMP, and varying NAD+. (C) NAD+ saturation curves of WT filaments treated with 0.5 mM ATP, 1 mM IMP, varying NAD+ and 2 mM GTP. (D) Six cryo-EM maps from two datasets (uninhibited ATP/IMP/NAD+ and partially inhibited ATP/IMP/NAD+/[2 mM]GTP) exhibiting a range of Bateman domain conformations. The uninhibited filaments were prepared with 0.5 mM ATP, 2 mM IMP, and 2 mM NAD+. The partially inhibited filaments were prepared with 0.5 mM ATP, 2 mM IMP, and 2 mM NAD+, and 2 mM GTP. (E) A view of a single tetramer from the inside of each octamer. The lighter colored tetramer from panel A is hidden, with the surface area buried between tetramer active sites colored in yellow, with the indicated total buried surface area. (F) Corresponding views of Bateman domain conformations. Protein displayed as ribbon, with the two interacting Bateman domains colored orchid and light pink. Cryo-EM density for non-protein ligand densities is colored green and red, for ATP and GTP, respectively. Symmetry identities labeled with gray letters. In the extended conformation, allosteric site 3 is distorted, and does not bind ligands (black dashes). Allosteric site 3 is formed in compressed protomers, but in the absence of guanine nucleotides it remains unoccupied (black asterisk). In the extended protomers of some bent octamers, is is possbile that allosteric site 2 is only partially occupied (black tilde).