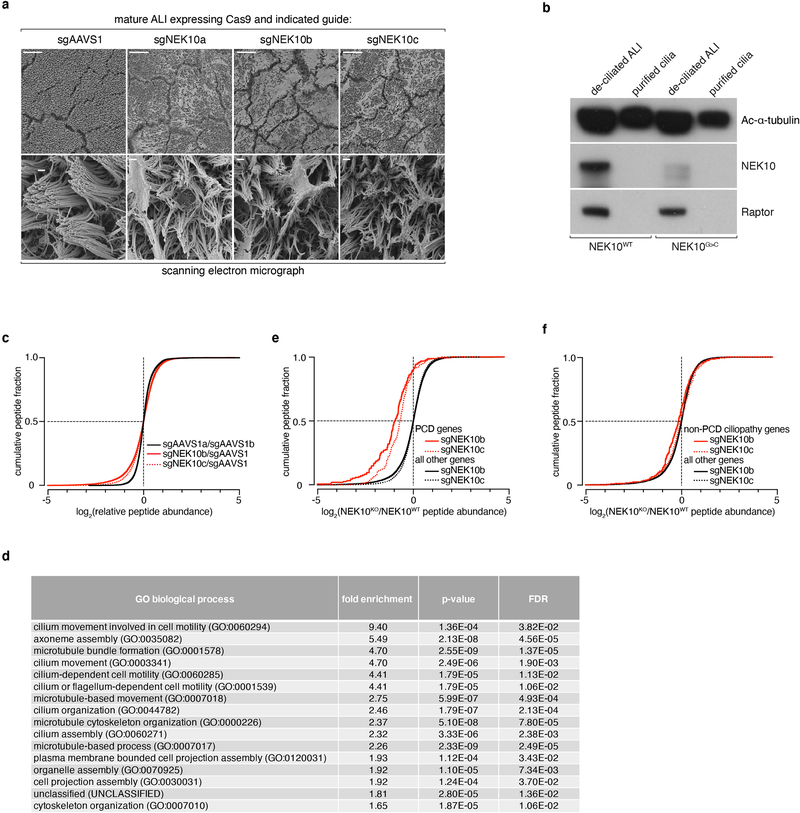
Extended Data Fig. 5: Structural and proteomic abnormalities in NEK10-deficient airway epithelium.
a, SEM of mature ALI edited with the indicated sgRNAs, scale bars 100μm (upper panels) and 1μm (lower panels), representative of 2 independent ALI differentiations. b, immunoblotting against the indicated proteins from lysates generated from purified cilia (lanes 2, 4) or remaining de-ciliated mature ALI (lanes 1, 3), representative of 2 experiments. c, cumulative distribution of phosphopeptides by log2 fold change between indicated conditions, solid (sgNEK10b) and dashed (sgNEK10c) red lines illustrate population of depleted phosphopeptides upon NEK10 deletion. d, table of gene ontology classes enriched among genes (n=395) whose peptides are depleted >1.5 fold (log2) after targeting with sgNEK10b, enrichment level, p-values, and false discovery rates (FDR) indicated. e, cumulative distribution of phosphopeptides by log2 fold change, previously validated PCD in red and all other detected proteins in black, as in (4e). f, cumulative distribution of phosphopeptides by log2 fold change, previously validated non-PCD ciliopathy loci in red and all other detected proteins in black, as in (4e).