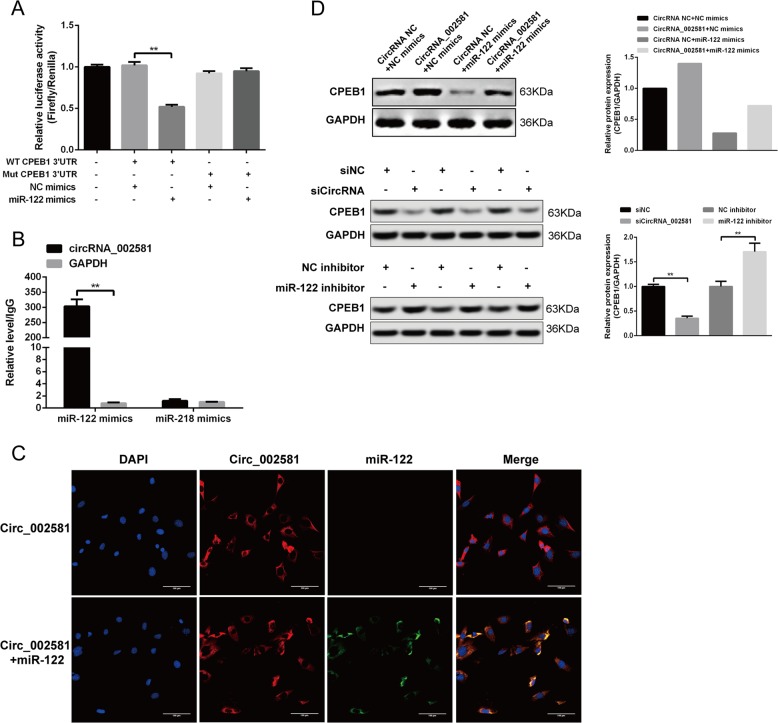
Fig. 2. Confirming the existence of circRNA_002581–miR-122–CPEB1 axis.
a Dual luciferase reporter gene analysis showed the direct binding between miR-122 and the 3′UTR wild type of CPEB1. The miRNA negative control mimics and the 3′UTR mutant type of CPEB1 have lost their binding capacity for each other. b RIP result showed circRNA_002581 in NIH3T3 cell lysis was pulled down and detected by qPCR with selected primer, indicating the existence of binding between circRNA_002581 and miR-122, where miR-218 was used as negative miRNA control and GAPDH was applied as control. c RNA FISH showed the co-localization of miR-122 (green) and circRNA_002581 (red) in the cytoplasm of HEK-293T cells. The blue spot is the nucleus dyed by DAPI (scale bar: 100 μm). d Western blot showed CPEB1 level was decreased when transfecting HEK-293T cells with miR-122 mimics than with NC mimics. Further co-transfecting circRNA_002581 could antagonize the inhibitory effect of miR-122 mimics on CPEB1 protein level (upper panel); CPEB1 protein level was significantly downregulated when transfected with siCircRNA_002581 in NCTC-1469 cells but significantly upregulated when transfected with miR-122 inhibitor (middle and lower panels). Error bars represent the SD. **p < 0.01.