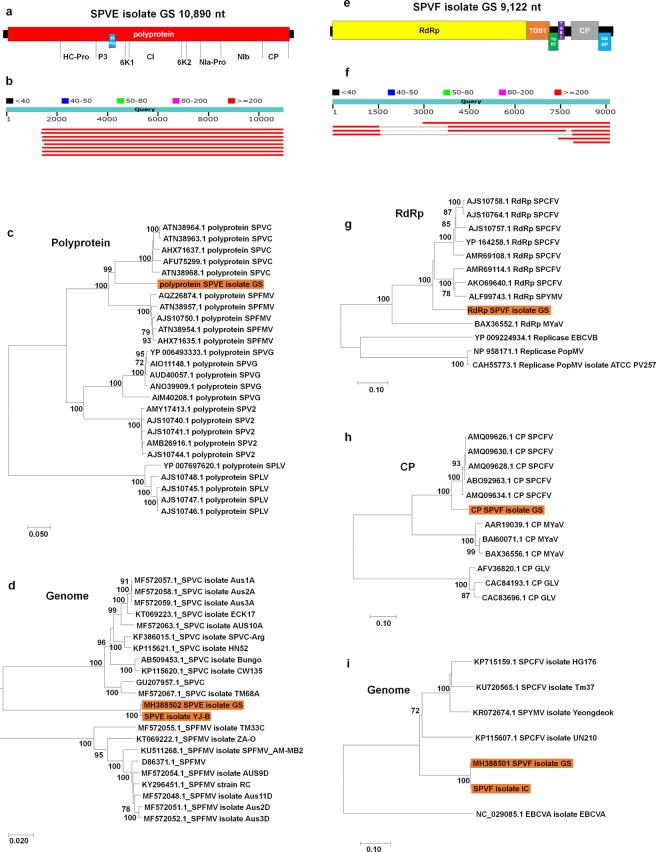
Figure 3.
Genome organization and phylogenetic analyses of two novel viruses, SPVE and SPVF. (a) Genome organization of SPVE isolate GS. Polyprotein cleavage sites were also indicated with respective protein names. (b) BLASTN results using complete genome of SPVE against NCBI’s NT database. (c) Maximum likelihood phylogenetic tree of polyprotein amino acid sequences for SPVC, SPVE, SPFMV, SPVG, SPV2, and SPLV. (d) Maximum likelihood phylogenetic tree of genome sequences for SPVC, SPFMV, SPVE, and SPFMV. Two genome sequences for SPVE isolates GS and YJ-B were included in the phylogenetic construction. (e) Genome organization of SPVF isolate GS. (f) BLASTN results using complete genome of SPVF against NCBI’s NT database. (g) Maximum likelihood phylogenetic tree of RdRp amino acid sequences for SPCFV, SPYMV, SPVF, Melon yellowing-associated virus (MYaV), Elderberry carlavirus B (EBCVB), and Poplar mosaic virus (PopMV). (h) Maximum likelihood phylogenetic tree of CP amino acid sequences for SPCFV, SPVF, MYaV, and Garlic latent virus (GLV). (i) Maximum likelihood phylogenetic tree of genome sequences for SPCFV, SPYMV, and SPVF. EBCVA was used as an outgroup. Two genome sequences for SPVF isolates GS and IC were included in the phylogenetic construction. For the phylogenetic tree construction, all available protein or genome sequences homologous to SPVE or SPVF were retrieved from GenBank based on BLASTP and BLASTN searches, respectively. Accession number, isolate name, and virus name were described. Orange color indicates SPVE or SPVF. We used bootstrap replication values of 1,000, and bootstrap values over 70% are shown.