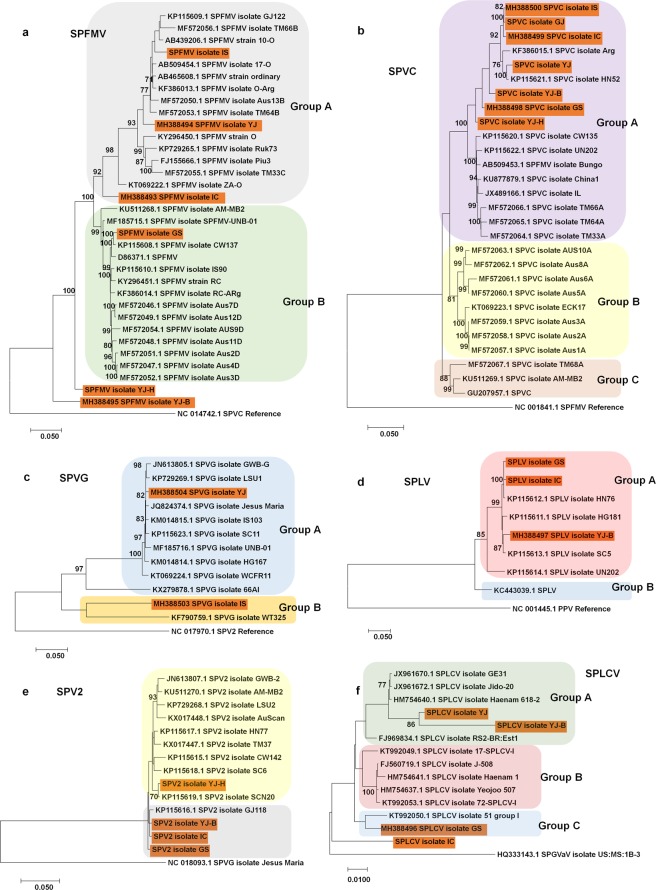
Figure 4.
Phylogenetic analyses of SPFMV, SPVC, SPVG, SPLV, SPV2, and SPLCV isolates. (a) Maximum likelihood phylogenetic tree of genome sequences for 33 SPFMV isolates including six isolates from this study indicated by orange color. SPVC was used as an outgroup. (b) Maximum likelihood phylogenetic tree of genome sequences for 28 SPVC isolates including seven isolates in this study indicated by orange color. SPFMV was used as an outgroup. (c) Maximum likelihood phylogenetic tree of genome sequences for 12 SPVG isolates including two isolates in this study indicated by orange color. SPV2 was used as an outgroup. (d) Maximum likelihood phylogenetic tree of genome sequences for eight SPLV isolates including three isolates in this study indicated by orange color. Plum pox virus (PPV) was used as an outgroup. (e) Maximum likelihood phylogenetic tree of genome sequences for 14 SPV2 isolates including four isolates in this study indicated by orange color. SPVG was used as an outgroup. (f) Maximum likelihood phylogenetic tree of genome sequences for 14 SPLCV isolates including four isolates in this study indicated by orange color. SPGVaV was used as an outgroup. For phylogenetic analyses, we retrieved only complete genome sequences for each virus from GenBank based on a BLASTN search. We used not only complete viral genome sequences with respective accession numbers but also nearly complete viral genome sequences (Table S2) without accession numbers in this study. Accession number, isolate name, and virus name were described. Orange color indicates virus genomes obtained from this study. We used bootstrap replication values of 1,000, and bootstrap values over 70% are shown.