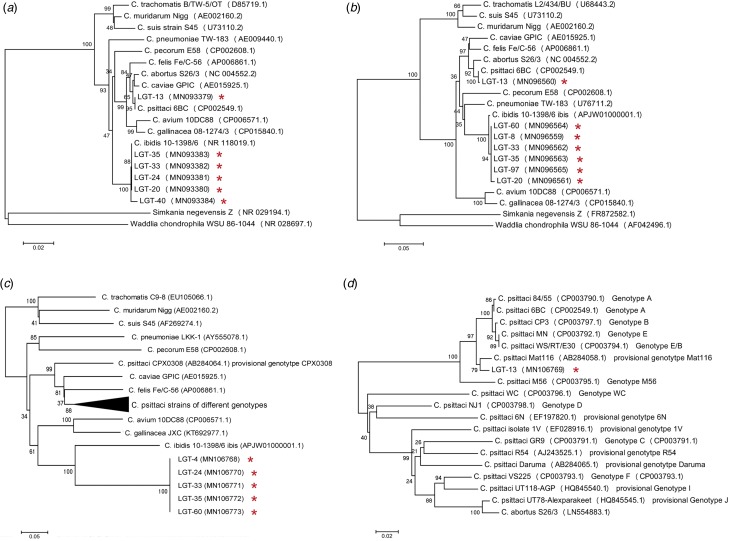
Fig. 1.
Phylogenetic tree based on 16S rRNA gene fragment (1356 bp) (a), 16S-23S intergenic spacer and full length of 23S rRNA domain I fragment (923 bp) (b) and ompA gene fragment (720 bp) of all Chlamydiaceae members (c) and C. psittaci strains of different ompA genotypes (d). Representative sequences of established Chlamydiaceae species as well as the sequences obtained from this study (GenBank accession number was marked by red asterisks) were included. S. negevensis strain Z and Waddlia chondrophila strain WSU 86-1044 were used as an outgroup. Based on these alignments, phylogenetic trees were constructed by the neighbour-joining method using the Maximum Composite Likelihood model with MEGA 5.05.