Abstract
Background
Women with epithelial ovarian cancer (OC) have a higher chance to benefit from poly (ADP-ribose) polymerase inhibitor (PARPi) therapy if their tumor has a somatic or hereditary BRCA1/2 pathogenic variant. Current guidelines advise BRCA1/2 genetic predisposition testing for all OC patients, though this does not detect somatic variants. We assessed the feasibility of a workflow for universal tumor DNA BRCA1/2 testing of all newly diagnosed OC patients as a prescreen for PARPi treatment and cancer predisposition testing.
Methods
Formalin-fixed paraffin-embedded tissue was obtained from OC patients in seven hospitals immediately after diagnosis or primary surgery. DNA was extracted, and universal tumor BRCA1/2 testing was then performed in a single site. Diagnostic yield, uptake, referral rates for genetic predisposition testing, and experiences of patients and gynecologists were evaluated.
Results
Tumor BRCA1/2 testing was performed for 315 (77.6%) of the 406 eligible OC samples, of which 305 (96.8%) were successful. In 51 of these patients, pathogenic variants were detected (16.7%). Most patients (88.2%) went on to have a genetic predisposition test. BRCA1/2 pathogenic variants were shown to be hereditary in 56.8% and somatic in 43.2% of patients. Participating gynecologists and patients were overwhelmingly positive about the workflow.
Conclusions
Universal tumor BRCA1/2 testing in all newly diagnosed OC patients is feasible, effective, and appreciated by patients and gynecologists. Because many variants cannot be detected in DNA from blood, testing tumor DNA as the first step can double the identification rate of patients who stand to benefit most from PARP inhibitors.
Hereditary BRCA1/2 pathogenic variants are present in 10%–20% of women with (nonborderline) epithelial ovarian, fallopian tube, and primary peritoneal cancer tumors (denoted as ovarian cancer [OC]). Early identification of pathogenic BRCA1/2 variants can reduce morbidity and mortality from breast cancer and OC by preventive and curative strategies in both patients and families (1,2). Generally, national guidelines therefore advise BRCA1/2 genetic predisposition testing for all OC patients (3,4). OC patients with pathogenic BRCA1/2 variants in their tumor are likely to benefit from poly (ADP-ribose) polymerase inhibitor (PARPi) therapy (5–8). PARPi therapy improves prognosis in patients with platinum-sensitive OC and is most effective in OC with defective homologous recombination mediated repair, especially BRCA1/2 defects (9–14). An estimated 18%–24% of patients with OC carry a BRCA1/2 pathogenic variant in their tumor, often in the context of an inherited germline BRCA1/2 variant. It is estimated that of the patients with a tumor BRCA1/2 variant, 54%–74% represent hereditary defects and 27%–46% somatic defects that are present in the tumor only (5–8).
Because a tumor BRCA1/2 test detects both hereditary and somatic BRCA1/2 variants, we developed a new workflow starting with universal tumor DNA BRCA1/2 testing in newly diagnosed OC patients. This workflow is based on our technical approach using single-molecule molecular inversion probe (smMIP) enrichment followed by next-generation sequencing (NGS) and allows rapid and reliable detection of both hereditary and somatic aberrations affecting BRCA1/2 in DNA derived from formalin-fixed paraffin-embedded (FFPE) tissue of OC (15).
The new universal tumor BRCA1/2 workflow has three key features. First, it detects many more patients who are eligible for PARPi therapy than conventional genetic predisposition testing of DNA from blood. Individuals can subsequently decide whether to continue with testing for a heritable BRCA1/2 variant. Second, testing tumor DNA effectively serves as a prescreen to tailor genetic counseling to those with a high a priori risk of a hereditary pathogenic variant. Third, by focusing on treatment options, the strategy may improve uptake of BRCA1/2 genetic predisposition in patients with OC.
In this study, the novel workflow of universal tumor DNA BRCA1/2 testing was implemented in seven hospitals. Its feasibility, effectiveness, and appreciation were evaluated by uptake, diagnostic yield, and referral rates for subsequent genetic counseling. The experiences of patients and gynecologists were systematically assessed.
Methods
Study Population
In this multicenter, observational cohort study, seven hospitals (two university and five regional) implemented universal tumor DNA BRCA1/2 testing in standard care between October 2015 and June 2017. Universal tumor BRCA1/2 testing was performed on tumor DNA from OC obtained by a diagnostic biopsy or a primary staging or cytoreductive surgery for OC. In a few cases, cells obtained from ascites aspiration were analyzed.
To measure uptake, OC patients (excluding borderline) were identified through a search of the nationwide registry of histo- and cytopathology (PALGA) (16). For all patients, referral for genetic counseling and genetic predisposition testing to the two genetic centers in the region and vital status were checked manually up to August 2018 (Supplementary Figure 1, available online). This study was approved by the privacy committee and the scientific board of PALGA and the medical ethical committee (NL 52165.091.15, 2015–1579). As required, written informed consent was obtained from the patients participating in the interviews. As part of diagnostics, patients provided oral informed consent for genetic testing.
Procedures
Pathologists were instructed to submit FFPE OC samples for centralized tumor DNA BRCA1/2 testing at the Radboud University Medical Center. Quality and quantity of tumor DNA (≥20% neoplastic cells) were evaluated centrally. Tumor DNA testing was performed with smMIP-based NGS of BRCA1/2, multiplex ligation-dependent probe amplification (MLPA), and methylation-specific MLPA of BRCA1 (15). Pathogenic and likely pathogenic variants (subsequently pathogenic variants) in BRCA1/2 were reported to the pathologist and gynecologist.
Gynecologists discussed the tumor DNA BRCA1/2 test and its results with the patient and were instructed to refer consenting patients with a tumor BRCA1/2 variant for genetic counseling if desired. Information for professionals and short videos for patients considering the difference between DNA testing in blood and in tumor tissue were provided after a positive tumor DNA BRCA1/2 test result and before genetic testing (see www.cancergenetics.eu).
Outcomes
The feasibility and effectiveness of the universal tumor BRCA1/2 workflow was assessed by its diagnostic yield, uptake of tumor DNA BRCA1/2 testing by pathologists, rate of successful testing, turnaround times, and referral rates for genetic counseling.
The appreciation of patients was assessed with semi-structured telephone interviews in a random sample of patients with somatic variants, germline variants, or without any variant. The appreciation of gynecologists in participating hospitals was assessed by a questionnaire.
Statistical Analysis
Patient groups were compared using Mann-Whitney U tests and Fisher exact tests. Bonferroni correction was applied when assessing differences between three patient groups. P values less than .05 (two-tailed) were considered statistically significant. Analyses were performed with R software (17).
Results
Uptake and Yield of Tumor DNA BRCA1/2 Testing
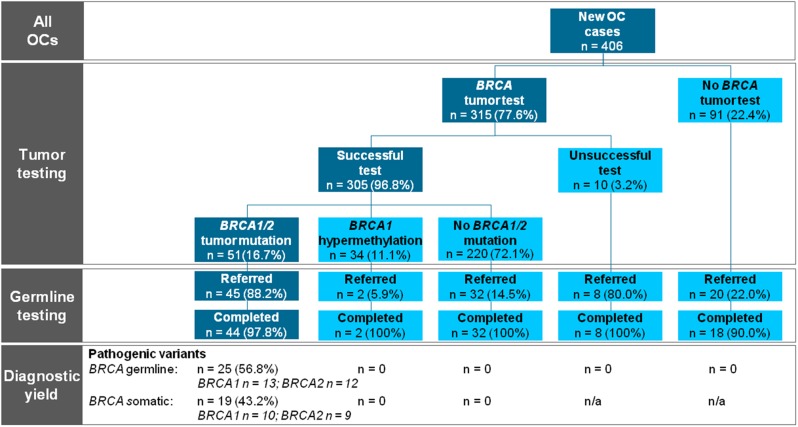
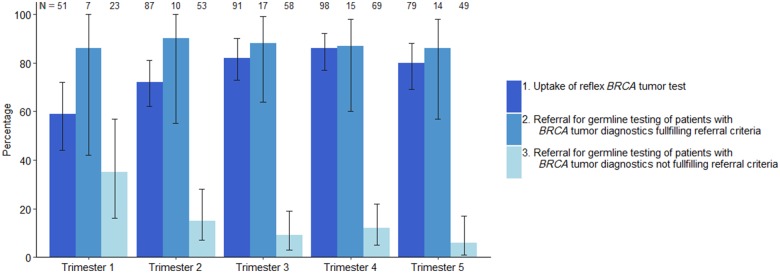
A BRCA1/2 tumor test was requested for 315 of 406 patients (77.6%) with newly diagnosed OC at a median age of 66 years (interquartile range [IQR] = 57–73; Table 1). Of the 91 patients without a tumor test, 20 patients (22.0%) were directly referred for genetic counseling and genetic predisposition testing (Figure 1). During the active implementation of universal tumor testing, uptake increased from 58.8% in the first 4 months of implementation to 79.7% in the last 4 months (Figure 2). The median time for tumor test request was 14 days (IQR = 8–21 days). Tumor BRCA1/2 smMIP-based NGS testing was successful for 305 of 315 patients (97%). MLPA was not possible in eight (3.1%) of 254 patients with a negative smMIP-based NGS test result.
Table 1.
Process evaluation of universal tumor BRCA1/2 testing in epithelial ovarian cancer patients
| Workflow characteristics | Total OC cohort n = 406 | BRCA1/2 tumor testing n = 315 |
|---|---|---|
| Age at OC diagnosis, median (IQR), y | 66 (57–73) | 66 (57–73) |
| Histology, No. (%) | ||
| Serous, low grade | 28 (6.9) | 18 (5.7) |
| Serous, high grade | 246 (60.6) | 199 (63.2) |
| Serous, unknown grade | 19 (4.7) | 19 (6.0) |
| Mucinous | 30 (7.4) | 17 (5.4) |
| Endometrioid | 23 (5.7) | 16 (5.1) |
| Clear cell | 25 (6.2) | 20 (6.3) |
| Carcinosarcoma | 13 (3.2) | 10 (3.2) |
| Adenocarcinoma NOS | 9 (2.2) | 3 (1.0) |
| Undifferentiated | 10 (2.5) | 10 (3.2) |
| Other | 3 (0.7) | 3 (1.0) |
| Uptake of tumor BRCA1/2 testing by pathologists, No. (%) | ||
| Overall uptake | 315 (78) | 315 (100) |
| Uptake within 3 months after OC diagnosis | 290 (71) | 290 (92) |
| Days from histological diagnosis to tumor BRCA1/2 test request, median (IQR) | – | 14 (8–2) |
| Outcome of BRCA1/2 tumor test, No. (%) | ||
| Successful test | – | 305 (96.8) |
| No test result available | – | 10 (3.2) |
| Insufficient quality of DNA material | – | 3 (1.0) |
| Low tumor cell percentage | – | 2 (0.6) |
| Lack of material | – | 4 (1.3) |
| Test retracted | – | 1 (0.3) |
| BRCA1/2 tumor tests needed, No. (%) | ||
| Successful after first test | – | 292 (95.7) |
| Successful after second test on different sample | – | 13 (4.3) |
| Tissue used for tumor BRCA1/2 test, No. (%) | ||
| Biopsy | – | 64 (20.3) |
| Ovariectomy, staging surgery, or debulking | – | 247 (78.4) |
| Ascites | – | 1 (0.3) |
| Other | – | 3 (1.0) |
| Tumor BRCA1/2 test on first histological sample, No. (%) | ||
| Yes | – | 209 (66.3) |
| No | – | 106 (33.7) |
| Chemotherapy before resection of tissue used for testing, No. (%) | ||
| Yes | – | 105 (33.3) |
| No | – | 209 (66.3) |
| Unknown | – | 1 (0.3) |
| BRCA1/2 tumor (likely) pathogenic variants, No. (%) | ||
| BRCA1 | – | 27 (8.9) |
| BRCA2 | – | 24 (7.9) |
| No (likely) pathogenic variant | – | 254 (83.3) |
| Days from BRCA1/2 tumor test request to test result, median (IQR) | 14 (12–16) | |
| Referral rates by indication status, No. (%) | ||
| Positive tumor BRCA1/2 test (n = 51) | – | 45 (88.2) |
| Unsuccessful tumor BRCA1/2 test (n =10) | – | 8 (80.0) |
| Negative tumor BRCA1/2 test (n = 254) | – | 34 (13.4) |
| ≥1 OC FDR of those referred (n = 34) | – | 2 (5.9) |
| No tumor BRCA1/2 test (n = 91) | – | 20 (22.0) |
| Appropriate referral rates | ||
| Referral of those with referral indication* (n = 63) | – | 55 (87.3) |
| No referral of those without referral indication (n = 252) | – | 220 (87.3) |
| Days from histological diagnosis to referral of those with positive tumor test (n = 45) | – | 68 (46–161) |
| Days from tumor BRCA1/2 test result to referral, median (IQR) | ||
| Referral time after any test result | – | 39 (19–126) |
| Referral time after test result indication to referral | – | 34 (22–102) |
Indication for referral for cancer predisposition testing is: a positive or unsuccessful test or a negative tumor test but a first-degree family history of OC. FDR = first-degree relative; IQR = interquartile range (ie, 25th–75th percentile); NOS = not otherwise specified; OC = epithelial ovarian cancer.
Figure 1.
Flowchart of the study showing the preferred universal tumor BRCA1/2 workflow on the left side in the dark boxes. n = 0 indicates that the test has been performed and no mutation was present. n = n/a indicates the test has not been performed or the test was unsuccessful and no result could be obtained. OC = epithelial ovarian cancer.
Figure 2.
Uptake of universal BRCA1/2 tumor testing and referral for genetic predisposition testing over time. Error bars represent 95% confidence intervals.
Fifty-one OC patients (16.7%, 95% confidence interval [CI] = 12.8% to 21.5%) had a tumor BRCA1/2 pathogenic variant, 27 (52.9%) in BRCA1 and 24 (47.1%) in BRCA2. Of 44 patients who completed genetic predisposition testing, 25 (56.8%, 95% CI = 41.1% to 71.3%) had hereditary pathogenic variants (Table 2), and 19 (43.2%, 95% CI = 28.7% to 58.9%) had somatic pathogenic variants (Table 3). BRCA1 hypermethylation was found in 34 of 305 cases (11.1%, 95% CI = 7.9% to 15.4%). Pathogenic variants and BRCA1 hypermethylation were mutually exclusive (Figure 1). Only two OC patients had BRCA variants of unknown significance in the absence of a pathogenic BRCA1 or BRCA2 variant (Supplementary Table 1, available online).
Table 2.
Characteristics of 25 epithelial ovarian cancer patients with a pathogenic BRCA1/2 hereditary variant identified with the universal tumor BRCA1/2 workflow
| Gene | Mutation c.* | Mutation p.* | Mutation position | Age OC, y | OC histology | FIGO stage | NCCN guideline | Personal history of breast cancer | FDR/SDR family history |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Breast cancer | OC | |||||||||
| BRCA1 | c.68_69del | p.(Glu23fs) | Neither | 50 | HGS | IIIc | Familial | – | FDR 48 | SDR 56 |
| BRCA1 | c.633C>T | p.(Gln172*) | Neither | 56 | HGS | IIIc | Personal | 47 ER- PR- Her2- | FDR 35 + 36, FDR 44, FDR 41 + 54, SDR 35, SDR 38, SDR 48 | SDR 61 |
| BRCA1 | c.981_982del | p.(Cys328*) | Neither | 55 | Undifferentiated | n/a | Personal | 53 TN | – | – |
| BRCA1 | c.2197_2201del | p.(Glu733fs) | OCCR | 57 | HGS | n/a | Familial | – | FDR 54 | FDR 53, FDR 84 |
| BRCA1 | c.2197_2201del | p.(Glu733fs) | OCCR | 73 | HGS | IIIc | No | – | FDR 70 | – |
| BRCA1 | c.2338C>T | p.(Gln780*) | OCCR | 41 | HGS | n/a | Familial | – | – | SDR 54 |
| BRCA1 | c.2685_2686del | p.(Pro897fs) | OCCR | 64 | HGS | n/a | Personal | 52 ER- PR- Her2? | SDR <40, SDR, SDR | – |
| BRCA1 | c.2722G>T | p.(Glu908*) | OCCR | 58 | HGS | n/a | Familial | – | FDR 41, SDR 65-70, SDR <43 | – |
| BRCA1 | c.2722G>T | p.(Glu908*) | OCCR | 67 | HGS | n/a | Personal | 53 TN | FDR 55 | – |
| BRCA1 | c.5095C>T | p.(Arg1699Trp) | Neither | 65 | HGS | n/a | Familial | – | FDR 36, SDR 75 | – |
| BRCA1 | c.5216A>T | p.(Asp1739Val) | Neither | 58 | HGS | n/a | No | – | FDR 57, SDR | – |
| BRCA1 | c.5291T>C | p.(Leu1764Pro) | BCCR | 42 | HGS | IV | No | – | – | – |
| BRCA1 | c.5309C>T | p.(Gly1770Val) | BCCR | 35 | HGS | Ic | No | – | – | – |
| BRCA2 | c.1147del | p.(Ile383fs) | BCCR | 49 | Clear cell | Ia | No | – | – | – |
| BRCA2 | c.3814del | p.(Met1272fs) | OCCR | 57 | HGS | n/a | No | 55 ER+ HR+ Her2? | – | – |
| BRCA2 | c.4314del | p.(Ala1439fs) | OCCR | 61 | Undifferentiated | II | No | – | – | – |
| BRCA2 | c.4449del | p.(Asp1484fs) | OCCR | 65 | Carcinosarcoma | n/a | No | – | – | – |
| BRCA2 | c.5213_5216del | p.(Thr1738fs) | OCCR | 51 | HGS | n/a | No | – | SDR 50 | – |
| BRCA2 | c.5213_5216del | p.(Thr1738fs) | OCCR | 55 | HGS | n/a | No | – | – | – |
| BRCA2 | c.5213_5216del | p.(Thr1738fs) | OCCR | 57 | HGS | IIIc | No | – | – | – |
| BRCA2 | c.5213_5216del | p.(Thr1738fs) | OCCR | 70 | HGS | n/a | Familial | – | FDR 44/52 | FDR 79 |
| BRCA2 | c.5645C>A | p.(Ser1882*) | OCCR | 52 | HGS | n/a | Familial | – | FDR 36 | – |
| BRCA2 | c.6275_6276del | p.(Leu2092fs) | Neither | 64 | HGS | n/a | Familial | 64 | FDR 45 | – |
| BRCA2 | c.7617 + 1G>T | p.? | Neither | 73 | HGS | n/a | Personal | 68 | FDR 35, FDR 57, FDR 65, SDR 40 | – |
| BRCA2 | c.9672dup | p.(Tyr322fs) | Neither | 79 | HGS | IIIc | No | 62 | – | – |
Variant nomenclature according to Human Genome Variation Society guidelines (varnomen.hgvs.org). FDR = first-degree relative; FIGO = International Federation of Gynecology and Obstetrics; HGS = high-grade serous; n/a = not available; NCCN = National Comprehensive Cancer Network; OC = epithelial ovarian cancer; SDR = second-degree relative; – = a negative history.
Table 3.
Characteristics of 19 epithelial ovarian cancer patients with a pathogenic somatic BRCA1/2 variant identified with the universal tumor BRCA1/2 workflow
| Gene | Mutation c.* | Mutation p.* | Age OC, y | OC histology | FIGO stage | NCCN guideline | Personal history of breast cancer | FDR or SDR family history |
|
|---|---|---|---|---|---|---|---|---|---|
| Breast cancer | OC | ||||||||
| BRCA1 | c.3013del | p.(Glu1005*) | 84 | HGS | n/a | No | – | – | – |
| BRCA1 | c.3031del | p.(Glu1011fs) | 55 | HGS | n/a | No | – | – | – |
| BRCA1 | c.3037del | p.(Glu1013fs) | 77 | HGS | n/a | No | – | – | – |
| BRCA1 | c.3205C>T | p.(Gln1069*) | 52 | Endometrioid | n/a | No | – | – | – |
| BRCA1 | c.3710dup | p.(Pro1238fs) | 59 | HGS | IIIc | Personal | 52 ER? HR? Her2? | SDR 68 | – |
| BRCA1 | c.4814T>A | p.(Leu1605*) | 77 | Undifferentiated | n/a | Personal | 54 ER? HR? Her2? | SDR 60 | – |
| BRCA1 | c.5363G>T | p.(Gly1788Val) | 68 | HGS | n/a | Familial | – | FDR 64 SDR 64 | – |
| BRCA1 | Duplication exons 1–20 | p.? | 41 | Mucineus | n/a | No | – | – | – |
| BRCA1 | Deletion of exons 1–11,or duplication of exons 11–24† | p.? | 53 | HGS | n/a | No | – | – | – |
| BRCA2 | c.3826G>T | p.(Glu1276*) | 46 | Endometrioid | n/a | No | 46 ER+ HR+ Her2– | – | – |
| BRCA2 | c.4085del | p.(His1362fs) | 54 | HGS | n/a | No | – | – | – |
| BRCA2 | c.4908del | p.(Val1637fs) | 76 | HGS | Ic | No | – | FDR 61 | – |
| BRCA2 | c.5048dup | p.(Thr1684fs) | 68 | HGS | IIIc | Personal | 65 ER+ PR+ Her2– | SDR 50 Male SDR age? | – |
| BRCA2 | c.5465del | p.(Asn1822fs) | 57 | HGS | IIIc | Familial | – | – | FDR 74 |
| BRCA2 | c.5942_5961del | p.(Ala1981fs) | 79 | HGS | IIIc | No | – | – | – |
| BRCA2 | c.7530_7531del | p.(Tyr2511fs) | 66 | Serous, grade unknown | n/a | No | – | SDR age? | – |
| BRCA2 | c.7792G>T | p.(Glu2598*) | 66 | HGS | n/a | No | – | FDR 70 | – |
| BRCA2 | c.799G>T | p.(Gly267*) | 49 | Endometrioid | IIIa | No | – | – | – |
| BRCA2 | c.9360del | p.(Ile3120fs) | 56 | Serous, low grade | Ic | No | – | – | – |
Variant nomenclature according to Human Genome Variation Society guidelines (varnomen.hgvs.org). FDR = first-degree relative; FIGO = International Federation of Gynecology and Obstetrics; HGS = high-grade serous; MLPA = multiplex ligation-dependent probe amplification; n/a = not available; NCCN = National Comprehensive Cancer Network; OC = epithelial ovarian cancer; SDR = second-degree relative; – = a negative history.
With MLPA, it is not possible to make a distinction between a 5’ deletion or 3’ duplication.
Patients with pathogenic hereditary BRCA1/2 variants were younger at OC diagnosis than patients without any tumor BRCA1/2 pathogenic variants (median 57 years, IQR = 52–65 years vs median 66 years, IQR = 57–73 years, respectively, P = .009) (Table 4; Supplementary Figure 2, available online). Patients with pathogenic hereditary BRCA1/2 variants were more likely to have high-grade serous OC than patients without tumor variants (84.0% vs 61.0%, P = .09), with no statistically significant difference between patients with a BRCA1 or BRCA2 variant (92.3% vs 75.0%, P = .26; Table 2). Based on their personal and family history of cancer, 13 (52.0%) patients with pathogenic hereditary BRCA1/2 variants (69.2% BRCA1 and 33.3% BRCA2, P = .12) already complied with National Comprehensive Cancer Network (NCCN) criteria for BRCA1/2 testing before their OC diagnosis (Table 2), suggesting their tumors could have been prevented by earlier genetic testing in relatives and presymptomatic cascade testing.
Table 4.
Characteristics of epithelial ovarian cancer patient by BRCA1/2 tumor and germline (hereditary) status
| Workflow characteristics | No BRCA1/2 mutation n = 254 | Germline mutation n = 25 | Somatic mutation *n = 19 |
P
†
|
||
|---|---|---|---|---|---|---|
| Germline vs somatic | Germline vs normal | Somatic vs normal | ||||
| Age at OC diagnosis, median (IQR), y | 66 (57–73) | 57 (52–65) | 59 (54–72) | 1.0 | .009 | .90 |
| Initial OC histology, No. (%) | .27 | 1.0 | 1.0 | |||
| Serous, low grade | 16 (6.3) | – | 1 (5.3) | |||
| Serous, high grade | 155 (61.0) | 21 (84.0) | 12 (63.2) | |||
| Serous, unknown grade | 17 (6.7) | – | 1 (5.3) | |||
| Mucinous | 16 (6.3) | – | 1 (5.3) | |||
| Endometrioid | 12 (4.7) | – | 3 (15.8) | |||
| Clear cell | 18 (7.1) | 1 (4.0) | – | |||
| Carcinosarcoma | 9 (3.5) | 1 (4.0) | – | |||
| Adenocarcinoma NOS | 2 (.8) | – | – | |||
| Undifferentiated | 6 (2.4) | 2 (8.0) | 1 (5.3) | |||
| HGS OC, No. (%) | ||||||
| Yes | 155 (61.0) | 21 (84.0) | 12 (63.2) | .49 | .09 | 1.0 |
| No | 99 (39.0) | 4 (16.0) | 7 (36.8) | |||
This includes only patients for whom the presence of a BRCA1/2 germline mutation was evaluated; hence, seven patients for whom results of germline diagnostics are unavailable were excluded. HGS = high-grade serous; IQR = interquartile range; NOS = not otherwise specified; OC = epithelial ovarian cancer; – = a count of zero.
P values were calculated with Mann-Whitney U tests or Fisher exact tests and adjusted with a Bonferroni correction for the three group comparisons.
Two tumors with a somatic pathogenic BRCA1/2 variant additionally harbored multiple other somatic variants in BRCA1 and/or BRCA2. In both tumors, a somatic pathogenic POLE variant was detected as the driver genetic defect.
Referral for Genetic Counseling and BRCA1/2 Genetic Predisposition Testing
Of 51 patients with a tumor BRCA1/2 pathogenic variant, 45 (88.2%) were referred for genetic counseling, of whom 44 chose genetic predisposition testing and 1 patient postponed her genetic counseling to a later time (Table 1). In total, six patients were not referred, of whom four died shortly after diagnosis. Two patients with a pathogenic BRCA1/2 variant in their OC were erroneously not referred. Of 254 patients without tumor BRCA1/2 pathogenic variants, 34 (13%) were referred for genetic counseling and genetic predisposition testing, including two referrals indicated by a first-degree relative with OC.
Of 315 patients with a tumor DNA BRCA1/2 test, 275 (87.3%) were either appropriately referred (55 of 63) or not referred (220 of 252) (Table 1). The appropriate referral rates remained stable over time, and the number of unjustified referrals—no tumor mutation and no suspected family history—decreased as fewer patients were referred before their negative tumor test result was available (Figure 2).
Patient Experiences
Of 18 invited patients, 13 (72.2%) consented to the telephone interview. This included four (30.8%) patients with hereditary, three (23.1%) with somatic, and six (46.2%) without BRCA1/2 pathogenic variants. Two patients (one with a somatic variant and one without a tumor BRCA1/2 pathogenic variant) were unaware of their tumor test. However, all appreciated that the tumor test was done, and they were satisfied with the amount and nature of the information provided by their physician. The patients specifically valued the speed with which the procedure allowed them to obtain relevant knowledge on heredity. Patients’ suggestions for improvement focused on reduction of waiting time after referral for genetic counseling to evaluate heredity of the identified tumor BRCA1/2 variant immediately.
Gynecologist Experiences
Questionnaire responses were received from 18 of 41 (43.9%) invited gynecologists. Fifteen (83.3%) were mostly or completely positive about universal tumor DNA BRCA1/2 testing, and one (5.6%) was mostly negative. Gynecologists indicated that the workflow could be further improved by quicker tumor test results (5 of 18), better communication of the tumor test results to the gynecologist (11 of 18), and more available information for patients (10 of 18). Sixteen gynecologists (88.9%) agreed with the statement that the gynecologist is the most suited physician to discuss tumor DNA BRCA1/2 testing, but one (5.6%) was of the opinion that tumor DNA BRCA1/2 testing should be performed only after formal clinical genetic counseling. The majority (14 of 18, 77.8%) of the gynecologists considered the test turnaround time to be adequate, but several gynecologists stated that they had to schedule an additional appointment to discuss the outcome. Explaining tumor DNA BRCA1/2 testing and reporting negative results took less than 5 minutes for all responding gynecologists, and one-half (9 of 18) needed more than 5 minutes to explain the relevance of a detected variant. The gynecologists perceived that the understanding of patients was considered good before testing in 55.6%. After disclosure and discussion of test results, this increased to 86.1%.
Discussion
Universal tumor DNA BRCA1/2 testing in OC patients was shown to be feasible in daily practice, and patients and gynecologists were overwhelmingly positive about this workflow. Universal tumor testing was performed in 77.6% of the patients and yielded 16.7% pathogenic BRCA1/2 variants in the tumor DNA, whereas a pathogenic variant would have been detected in only 9.5% of patients with universal genetic predisposition testing. Hence, the universal tumor BRCA1/2 workflow identified twice as many patients for PARPi therapy than conventional genetic predisposition testing of DNA from blood could have detected.
PARPi therapies are becoming more and more important to improve the generally poor prognosis of OC (14,18). Under current universal OC genetic predisposition testing guidelines, up to 90.5% of patients would receive two genetic tests because they are negative for a hereditary variant and would need a subsequent tumor test to identify somatic BRCA1/2 variants. The universal tumor testing workflow is more effective and reduces the number of tests by 38.7% because in about 16.7% of the patients, subsequent heredity genetic counseling and a blood test are needed. Furthermore, tumor BRCA1/2 status is available shortly after OC diagnosis, which is advantageous now that maintenance treatment with PARPi after first-line chemotherapy has been shown to be effective (14).
By focusing on treatment options, the strategy may improve uptake of BRCA1/2 testing in patients with OC. In our study, we showed that after an implementation learning period of 6 months, 70.4% of the OC followed the intended workflow: 81.6% of newly diagnosed OC patients received universal tumor DNA BRCA1/2 testing, and 86.3% of those who tested positive received genetic counseling and testing. The performance of the workflow can likely be generalized to each hospital setting because no difference existed between university and community hospitals. The high uptake may also be reached by conventional genetic predisposition testing because active implementation of universal testing yielded uptake rates up to 74%–90% (19–21), which is far beyond reported uptake of universal BRCA1/2 genetic predisposition testing in OC in the United States and Canada of only 10%–30% (22,23). However, the waiting lists for genetic counseling and genetic predisposition testing may stagnate the increasing uptake (24). Testing tumor DNA will effectively serve as a prescreen to tailor genetic counseling to those with a high a priori risk of a hereditary pathogenic variant. From our data, we conclude that it will diminish by sixfold the number of patients needing genetic counseling and genetic predisposition testing.
In this study, tumor BRCA1/2 pathogenic variants were identified in 16.7% of OCs. This rate is in line with previously reported rates of 18%–24% (6–8). The proportion of pathogenic hereditary BRCA1/2 variants of 56.8% is similar to rates found in another recent cohort (54%) but low compared with 62%–74% in other studies (5–8). Possible explanations can be low prevalence of founder mutations and reduction of BRCA1/2-associated tumors due to relatively high uptake of predictive genetic testing and subsequent preventive measures in our population (25).
It is essential that the test on FFPE tumor material as a prescreen for genetic counseling and genetic predisposition testing is equally reliable as genetic predisposition testing on DNA derived from peripheral blood lymphocytes. Our validated approach with smMIP-based NGS and MLPA detects all known pathogenic hereditary BRCA1/2 variants (15). During our study, the CNV analysis by MLPA was impossible for 3.1% OCs with successful smMIP-based NGS, which leads to a minor decrease in sensitivity of the tumor test. However, exon deletions and duplications form a minority of the pathogenic hereditary variants, and only two somatic exon deletions or duplications were present in the 297 samples that could be evaluated for these types of variants. Overall, we conclude that it is safe and feasible to replace BRCA1/2 genetic predisposition testing by BRCA1/2 tumor DNA testing as a prescreen for BRCA1/2 genetic predisposition testing when performed in an accredited genetic diagnostic laboratory after rigorous validation of the assay.
A strength of this study is that the novel workflow was implemented in the daily clinical practice of multiple centers through which realistic uptake rates could be assessed. However, this limited the possibility to assess why patients did not undergo a tumor test or predisposition testing. The uptake of cancer predisposition testing may therefore be even higher because some patients may have received testing at other genetic centers.
In conclusion, universal BRCA1/2 tumor testing in all new OC patients simultaneously facilitates treatment and genetic screening purposes and is very well appreciated by participating patients and gynecologists. It is more effective than current guideline advice with universal genetic predisposition testing because it reduces the number of tests and consultations needed and provides information on tumor BRCA1/2 variants shortly after diagnosis, enabling personalized treatment choices with PARPi. It also reaches patients with somatic variants and patients who are underrepresented in clinical genetic counseling and genetic predisposition testing, such as patients with a low educational level and non-Caucasian background (26). Universal tumor DNA BRCA1/2 testing is a robust and suitable option that effectively and efficiently increases the opportunity for targeted treatment in patients and cancer prevention in relatives.
Funding
This work was financially supported by AstraZeneca.
Notes
AstraZeneca was not involved in any aspect of the study, including design; collection, analysis, and interpretation of the data; writing of the manuscript; or decision to submit it for publication. AstraZeneca was informed in advance about the submission of the manuscript. The authors declare no conflicts of interest.
The OPA working group consists of the following members: Canisius Wilhelmina Ziekenhuis, Nijmegen, the Netherlands: Inge M. W. Ebisch, Ineke M. de Kievit, Nicole G. Laurens; Radboud University Medical Center, Nijmegen, the Netherlands: Nelleke Ottevanger; Elizabeth Tweesteden Ziekenhuis, Tilburg, the Netherlands: Robbert J. van Alphen, M. Caroline Vos, Anneke A. M. van den Wurff; Zuyderland ziekenhuis, Heerlen/Sittard, the Netherlands: Roel van Kampen, Helen J. M. M. Mertens, Prapto Sastrowijoto, Mirjam J. A. Engelen; Maastricht University Medical Center, Maastricht, the Netherlands: Roy I. Lalisang, Brigitte F. M. Slangen, Loes F. S. Kooreman; Catharina Ziekenhuis, Eindhoven, the Netherlands: Geert-Jan M. Creemers, Jurgen M. J. Piek; Stichting PAMM, Laboratory for Pathology, Eindhoven, the Netherlands: Paul Klinkhamer, Judith W. M. Jeuken; Rijnstate Ziekenhuis, Arnhem, the Netherlands: Anette L. Aalders, Riena P. Aliredjo.
We thank the Dutch Pathology Database (PALGA) and the study contact persons (Lucy I. H. Overbeek and Annette H. Gijsbers-Bruggink) for the retrieval of the pathology data and the contribution to this study.
Supplementary Material
Contributor Information
OPA Working Group:
Inge M W Ebisch, Ineke M de Kievit, Nicole G Laurens, Nelleke Ottevanger, Robbert J van Alphen, M Caroline Vos, Anneke A M van den Wurff, Roel van Kampen, Helen J M M Mertens, Prapto Sastrowijoto, Mirjam J A Engelen, Roy I Lalisang, Brigitte F M Slangen, Loes F S Kooreman, Geert-Jan M Creemers, Jurgen M J Piek, Paul Klinkhamer, Judith W M Jeuken, Anette L Aalders, and Riena P Aliredjo
References
- 1. Domchek SM, Friebel TM, Singer CF, et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010;304(9):967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Evans DG, Harkness EF, Howell A, et al. Intensive breast screening in BRCA2 mutation carriers is associated with reduced breast cancer specific and all cause mortality. Hered Cancer Clin Pract. 2016;14:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Comprehensive Cancer Network. Genetic/Familial High-Risk Assessment: Breast and Ovarian (Version I.2018 - October 3, 2017). 2018. https://www.nccn.org/professionals/physician_gls/PDF/genetics_screening.pdf. Accessed March 20, 2018.
- 4. Arts-de Jong M, de Bock GH, van Asperen CJ, Mourits MJ, de Hullu JA, Kets CM.. Germline BRCA1/2 mutation testing is indicated in every patient with epithelial ovarian cancer: a systematic review. Eur J Cancer. 2016;61:137–145. [DOI] [PubMed] [Google Scholar]
- 5. de Jonge MM, Ruano D, van Eijk R, et al. Validation and implementation of BRCA1/2 variant screening in ovarian tumor tissue. J Mol Diagn. 2018;20(5):600–611. [DOI] [PubMed] [Google Scholar]
- 6. Hennessy BT, Timms KM, Carey MS, et al. Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol. 2010;28(22):3570–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20(3):764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. [DOI] [PubMed] [Google Scholar]
- 10. Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375(22):2154–2164. [DOI] [PubMed] [Google Scholar]
- 11. Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390(10106):1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ledermann JA, Harter P, Gourley C, et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. 2016;17(11):1579–1589. [DOI] [PubMed] [Google Scholar]
- 13. Pujade-Lauraine E, Ledermann JA, Selle F, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(9):1274–1284. [DOI] [PubMed] [Google Scholar]
- 14. Moore K, Colombo N, Scambia G, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379(26):2495–2505. [DOI] [PubMed] [Google Scholar]
- 15. Weren RD, Mensenkamp AR, Simons M, et al. Novel BRCA1 and BRCA2 tumor test as basis for treatment decisions and referral for genetic counselling of patients with ovarian carcinomas. Hum Mutat. 2017;38(2):226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Casparie M, Tiebosch AT, Burger G, et al. Pathology databanking and biobanking in the Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell Oncol. 2007;29(1):19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Core Team. R: A Language and Environment for Statistical Computing [Computer Program]. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 18. Ledermann JA, Harter P, Gourley C, et al. Quality of life during olaparib maintenance therapy in platinum-sensitive relapsed serous ovarian cancer. Br J Cancer. 2016;115(11):1313–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swanson CL, Kumar A, Maharaj JM, et al. Increasing genetic counseling referral rates through bundled interventions after ovarian cancer diagnosis. Gynecol Oncol. 2018;149(1):121–126. [DOI] [PubMed] [Google Scholar]
- 20. Uyar D, Neary J, Monroe A, Nugent M, Simpson P, Geurts JL.. Implementation of a quality improvement project for universal genetic testing in women with ovarian cancer. Gynecol Oncol. 2018;149(3):565–569. [DOI] [PubMed] [Google Scholar]
- 21. Kentwell M, Dow E, Antill Y, et al. Mainstreaming cancer genetics: a model integrating germline BRCA testing into routine ovarian cancer clinics. Gynecol Oncol. 2017;145(1):130–136. [DOI] [PubMed] [Google Scholar]
- 22. Childers CP, Childers KK, Maggard-Gibbons M, Macinko J.. National estimates of genetic testing in women with a history of breast or ovarian cancer. J Clin Oncol. 2017;35(34):3800–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCuaig JM, Armel SR, Care M, Volenik A, Kim RH, Metcalfe KA.. Next-generation service delivery: a scoping review of patient outcomes associated with alternative models of genetic counseling and genetic testing for hereditary cancer. Cancers. 2018;10(11): 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCuaig JM, Stockley TL, Shaw P, et al. Evolution of genetic assessment for BRCA-associated gynaecologic malignancies: a Canadian multisociety roadmap. J Med Genet. 2018;55(9):571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harmsen MG, Arts-de Jong M, Horstik K, et al. Very high uptake of risk-reducing salpingo-oophorectomy in BRCA1/2 mutation carriers: a single-center experience. Gynecol Oncol. 2016;143(1):113–119. [DOI] [PubMed] [Google Scholar]
- 26. van der Giessen JAM, van Riel E, Velthuizen ME, van Dulmen AM, Ausems M.. Referral to cancer genetic counseling: do migrant status and patients' educational background matter? J Community Genet. 2017;8(4):303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.